Search Count: 39
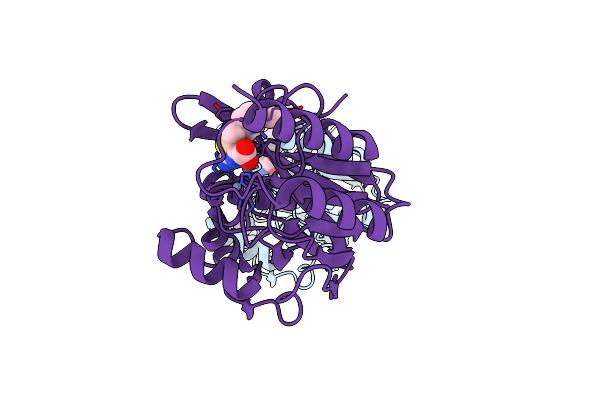 |
Crystal Structure Of B1 Vim-2 Mbl In Complex With 2-Amino-5-(4-Methoxybenzyl)Thiazole-4-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 4YJ |
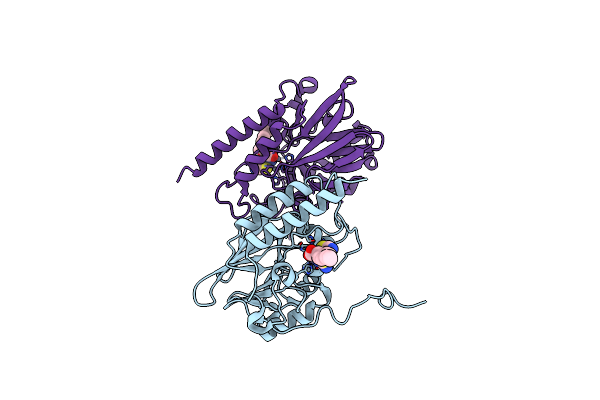 |
Crystal Structure Of B3 L1 Mbl In Complex With 2-Amino-5-(4-Propylbenzyl)Thiazole-4-Carboxylic Acid
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 51I, SCN |
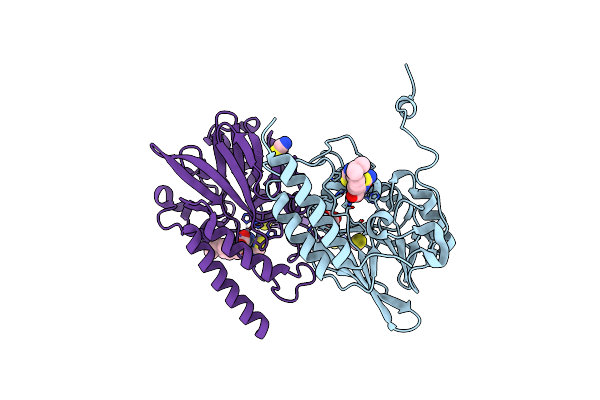 |
Crystal Structure Of B3 L1 Mbl In Complex With 2-Amino-5-(4-Isopropylbenzyl)Thiazole-4-Carboxylic Acid
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5A5, SCN, GOL |
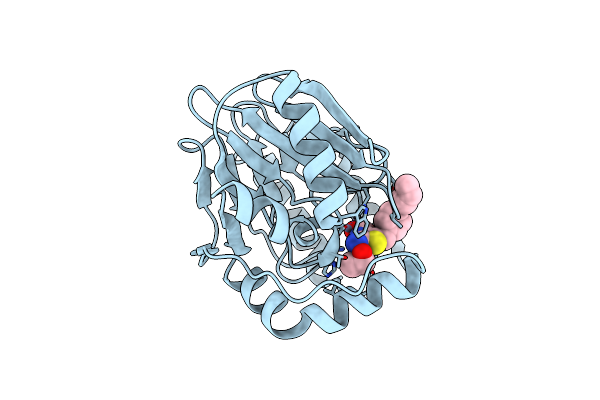 |
Crystal Structure Of B2 Sfh-I Mbl In Complex With 2-Amino-5-(4-(But-3-En-1-Yloxy)Benzyl)Thiazole-4-Carboxylic Acid
Organism: Serratia fonticola
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5I6, EOH |
 |
Crystal Structure Of B1 Vim-2 Mbl In Complex With 2-Amino-5-Isobutylthiazole-4-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5IY, GOL |
 |
Crystal Structure Of B1 Vim-2 Mbl In Complex With 2-Amino-5-(But-3-En-1-Yl)Thiazole-4-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5YT |
 |
Crystal Structure Of B1 Vim-2 Mbl In Complex With 2-Amino-5-Pentylthiazole-4-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5Z2, FMT |
 |
Crystal Structure Of B1 Vim-2 Mbl In Complex With 2-Amino-5-Hexylthiazole-4-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5ZD |
 |
Crystal Structure Of B1 Vim-2 Mbl In Complex With 2-Amino-5-Heptylthiazole-4-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5ZR, FMT |
 |
Crystal Structure Of B1 Vim-2 Mbl In Complex With 2-Amino-5-(Thiophen-2-Ylmethyl)Thiazole-4-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5ZU |
 |
Crystal Structure Of B1 Vim-2 Mbl In Complex With 2-Amino-5-Phenethylthiazole-4-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5ZX, FMT |
 |
Crystal Structure Of B1 Ndm-1 Mbl In Complex With 2-Amino-5-Phenethylthiazole-4-Carboxylic Acid
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5ZX, GOL |
 |
Crystal Structure Of B1 Vim-2 Mbl In Complex With 2-Amino-5-(2-(Thiophen-2-Yl)Ethyl)Thiazole-4-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 60J |
 |
Crystal Structure Of B1 Imp-1 Mbl In Complex With 2-Amino-5-Phenethylthiazole-4-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: ZN, 5ZX |
 |
Crystal Structure Of Vim-2 Mbl In Complex With 1-((2-Aminobenzo[D]Thiazol-6-Yl)Methyl)-1H-Imidazole-2-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-01-26 Classification: HYDROLASE Ligands: ZN, HQF, PEG |
 |
Crystal Structure Of Vim-2 Mbl In Complex With 1-(But-3-En-1-Yl)-4-Methyl-1H-Imidazole-2-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-01-26 Classification: HYDROLASE Ligands: ZN, HQR, GOL |
 |
Crystal Structure Of Vim-2 Mbl In Complex With 1-(But-3-En-1-Yl)-5-Methyl-1H-Imidazole-2-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.23 Å Release Date: 2022-01-26 Classification: HYDROLASE Ligands: ZN, HQ0 |
 |
Crystal Structure Of Vim-2 Mbl In Complex With 1-Benzyl-5-Methyl-1H-Imidazole-2-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2022-01-26 Classification: HYDROLASE Ligands: ZN, HQ3 |
 |
Crystal Structure Of Vim-2 Mbl In Complex With 1-Propyl-1H-Imidazole-2-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2022-01-19 Classification: HYDROLASE Ligands: ZN, HKR, FMT |
 |
Crystal Structure Of Vim-2 Mbl In Complex With 1-Isopropyl-1H-Imidazole-2-Carboxylic Acid
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-01-19 Classification: HYDROLASE Ligands: ZN, HKU, FMT |

