Search Count: 14
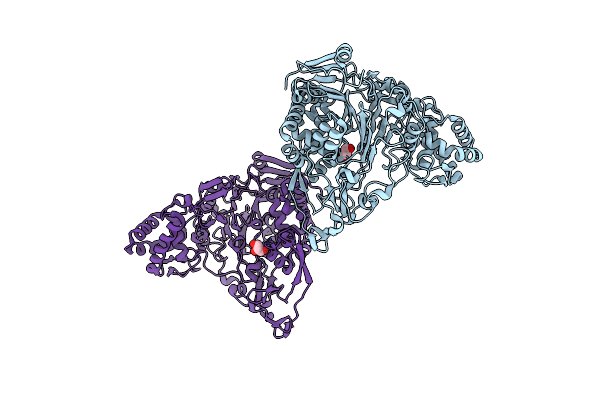 |
Crystal Structures Of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight Into Autoproteolytic Activation
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-07-06 Classification: HYDROLASE Ligands: GOL |
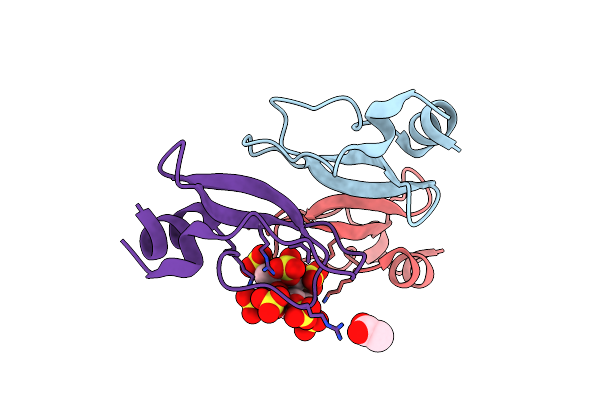 |
Crystal Structure Of Aprotinin In Complex With Sucrose Octasulfate: Unusual Interactions And Implication For Heparin Binding
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-09-15 Classification: HYDROLASE INHIBITOR Ligands: ACT |
 |
Crystal Structure Of Aprotinin In Complex With Sucrose Octasulfate: Unusual Interactions And Implication For Heparin Binding
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-06-23 Classification: HYDROLASE INHIBITOR Ligands: SO4, HG, GOL |
 |
Crystal Structure Of Aprotinin In Complex With Sucrose Octasulfate: Unusual Interactions And Implication For Heparin Binding
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-06-23 Classification: HYDROLASE INHIBITOR |
 |
Structural Basis For The Iron Uptake Mechanism Of Helicobacter Pylori Ferritin
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-01-13 Classification: OXIDOREDUCTASE Ligands: GOL |
 |
Structural Basis For The Iron Uptake Mechanism Of Helicobacter Pylori Ferritin
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-01-13 Classification: OXIDOREDUCTASE Ligands: FE, IPA, GOL |
 |
Structural Basis For The Iron Uptake Mechanism Of Helicobacter Pylori Ferritin
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-01-13 Classification: OXIDOREDUCTASE Ligands: FE, GOL |
 |
Structural Basis For The Iron Uptake Mechanism Of Helicobacter Pylori Ferritin
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-01-13 Classification: OXIDOREDUCTASE Ligands: FE, GOL |
 |
Structural Basis For The Iron Uptake Mechanism Of Helicobacter Pylori Ferritin
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-01-13 Classification: OXIDOREDUCTASE Ligands: FE, GOL |
 |
Crystal Structures Of Glutaryl 7-Aminocephalosporanic Acid Acylase: Mutational Study Of Activation Mechanism
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2006-01-24 Classification: HYDROLASE |
 |
Glutaryl 7-Aminocephalosporanic Acid Acylase: Mutational Study Of Activation Mechanism
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-01-24 Classification: HYDROLASE Ligands: GOL |
 |
Glutaryl 7-Aminocephalosporanic Acid Acylase: Mutational Study Of Activation Mechanism
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-01-24 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Glutaryl 7-Aminocephalosporanic Acid Acylase: Mutational Study Of Activation Mechanism
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2006-01-24 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Crystal Structures Of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight Into Autoproteolytic Activation
Organism: Pseudomonas sp. sy-77-1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-03-11 Classification: HYDROLASE Ligands: EDO |

