Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
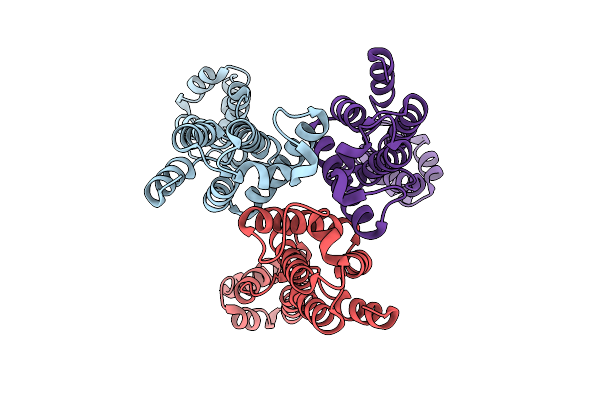 |
Organism: Human t-cell leukemia virus type i
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN |
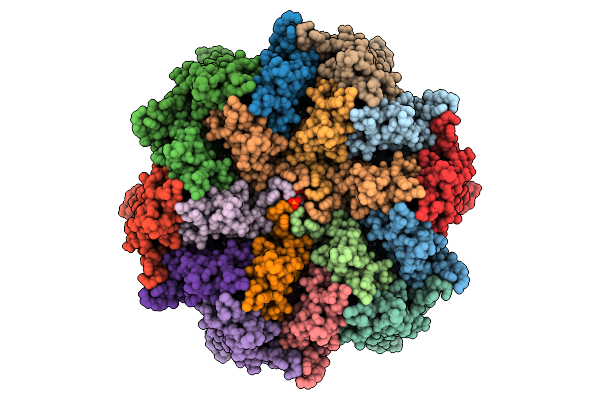 |
High-Resolution Analysis Of The Human T-Cell Leukemia Virus Capsid Protein Reveals Insights Into Immature Particle Morphology
Organism: Human t-cell leukemia virus type i
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRUS LIKE PARTICLE Ligands: IHP |
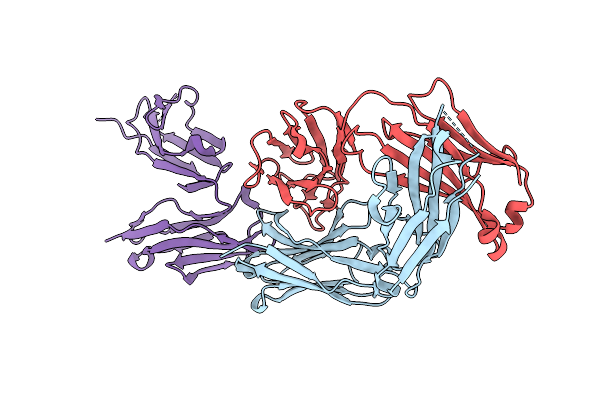 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA |
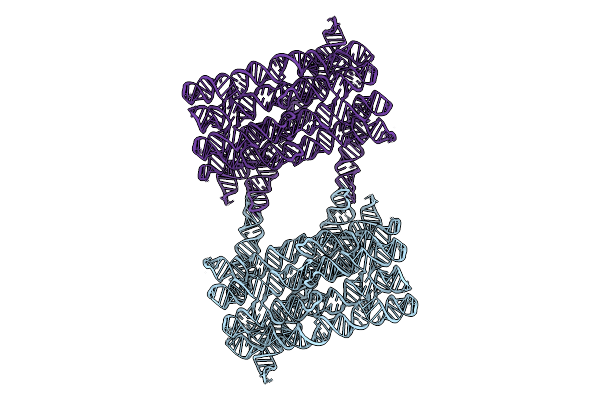 |
1-Methyl-Pseudouridine Twist Corrected Rna Origami 6-Helix Bundle Type-1 Dimer
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA |
 |
Organism: Tetrahymena thermophila
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
 |
1-Methyl-Pseudouridine L-21 Scai Tetrahymena Ribozyme - Extended Conformation
Organism: Tetrahymena thermophila
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
 |
Organism: Tetrahymena thermophila
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
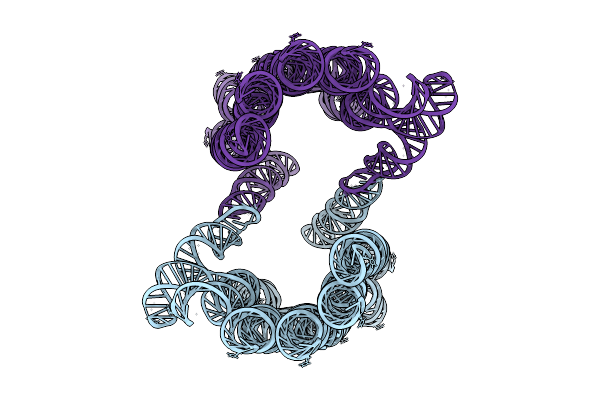 |
1-Methyl-Pseudouridine Twist Corrected Rna Origami 6-Helix Bundle Type-2 Dimer
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RNA |
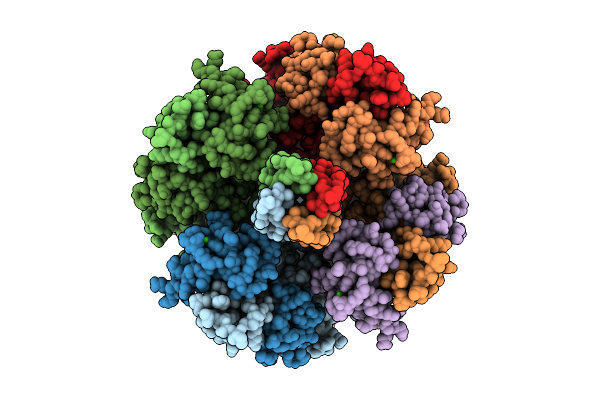 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PT5, K, CA |
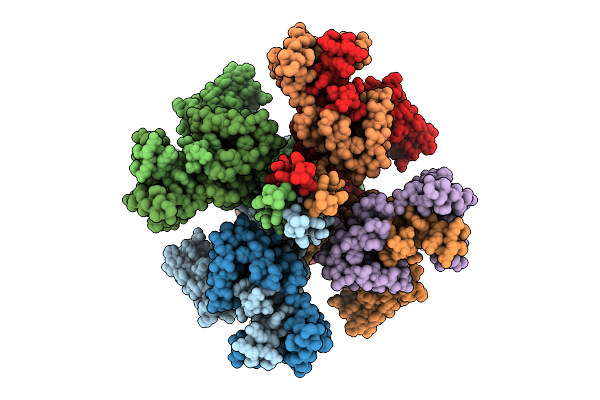 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PIO, K |
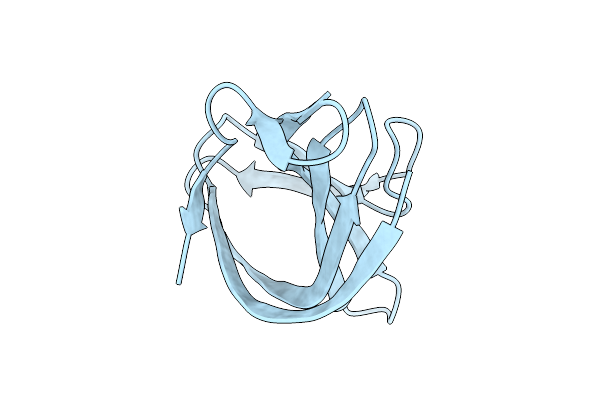 |
Organism: Streptococcus phage philp081102
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LIGASE |
 |
Cryo-Em Structure Of Slc30A10 In Mn2+-Bound State, Determined In Inward-Facing Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN Ligands: MN |
 |
Cryo-Em Structure Of Slc30A10, Determined In Asymmetric Conformations-One Subunit In An Inward-Facing Mn2+-Bound And The Other In An Outward-Facing Mn2+-Unbound Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN Ligands: MN |
 |
Cryo-Em Structure Of Slc30A10 In The Absence Of Mn2+, Determined In Inward-Facing Conformation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: HVG, A1ENS |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: MEMBRANE PROTEIN Ligands: GLU |