Search Count: 412
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: SO4, A1L38 |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Ossps3 Complexed With Zoledronate And Isopentenyl Diphosphate
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: IPE, ZOL, MG |
 |
Organism: Oryza sativa subsp. japonica
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of The Sps3-Fbn5 Complex From Oryza Sativa In Complex With Cobalt And Geranylgeranyl S-Thiodiphosphate (Ggspp)
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: GGS, CO |
 |
Organism: Homo sapiens, Oplophorus gracilirostris, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: HOH |
 |
Organism: Mus musculus, Homo sapiens, Bos taurus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus, Homo sapiens, Bos taurus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: CO, 92X |
 |
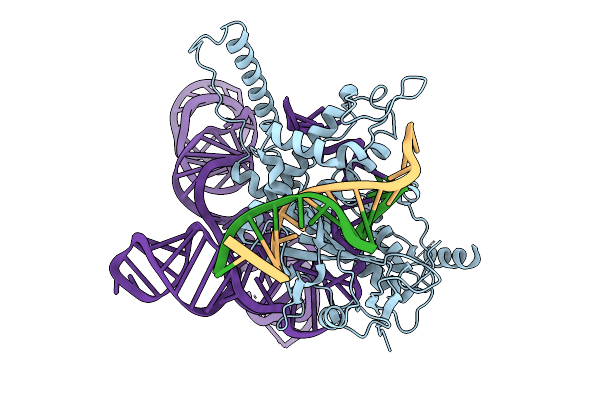
Structure Of Latranc Complex Bound To 27Nt Complementary Dna Substrate, Conformation 1
Organism: Lawsonibacter sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: IMMUNE SYSTEM/DNA/RNA |
 |
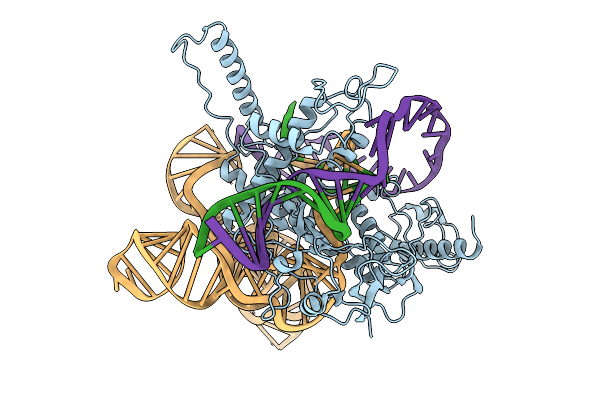
Organism: Lawsonibacter sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: IMMUNE SYSTEM/DNA/RNA |
 |
Structure Of Latranc Complex Bound To 27Nt Complementary Dna Substrate, Conformation 2
Organism: Lawsonibacter sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: IMMUNE SYSTEM/DNA/RNA |
 |
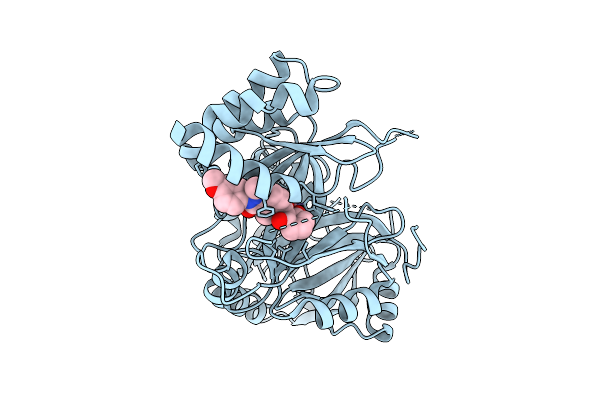
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: A1L4S, CO |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: OXIDOREDUCTASE Ligands: A1L4A, CO |
 |
Organism: Orthobornavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: TRANSFERASE, VIRAL PROTEIN Ligands: ZN |
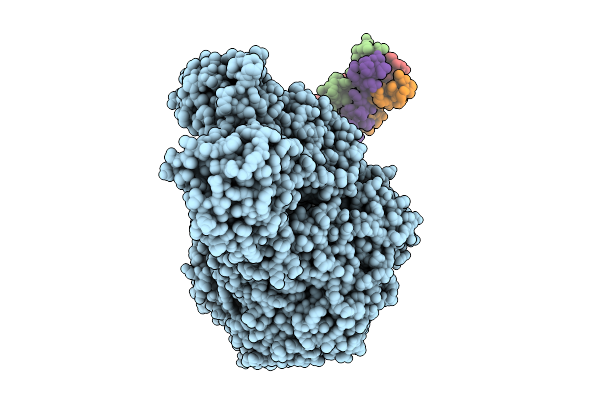 |
Organism: Orthobornavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: TRANSFERASE, VIRAL PROTEIN Ligands: ZN |
 |
Organism: Orthobornavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: TRANSFERASE, VIRAL PROTEIN Ligands: ZN |
 |
Organism: Orthobornavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: TRANSFERASE, VIRAL PROTEIN Ligands: ZN |

