Search Count: 23
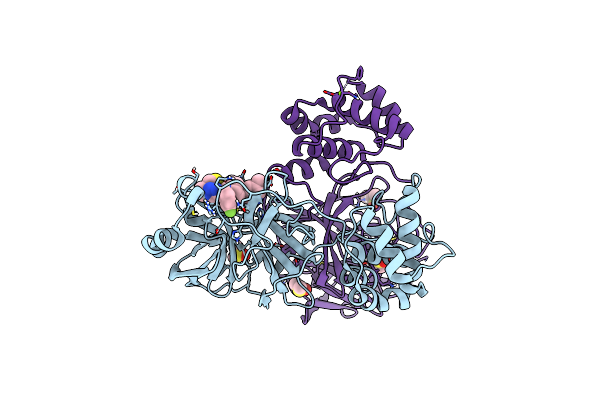 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Noncovalently Bound Inhibitor C5N17A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: DMS, A1IHT, CL, MG |
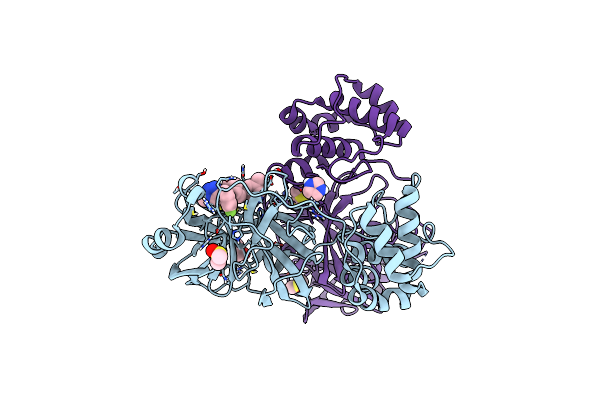 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Noncovalently Bound Inhibitor C5N17B
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: DMS, A1IHV, IMD |
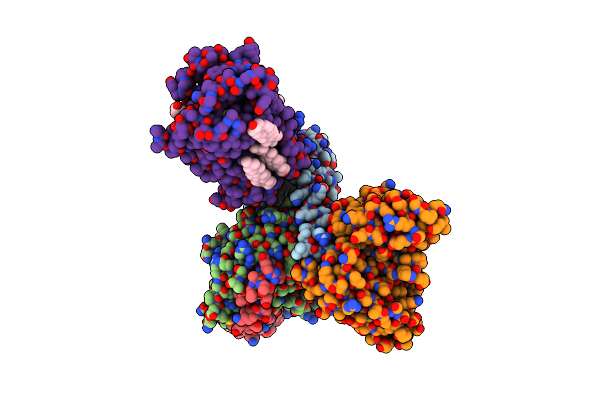 |
Cryo-Em Structure Of Histamine H3 Receptor In Complex With Immethridine And Minigo
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: SIGNALING PROTEIN Ligands: A1LY2, CLR |
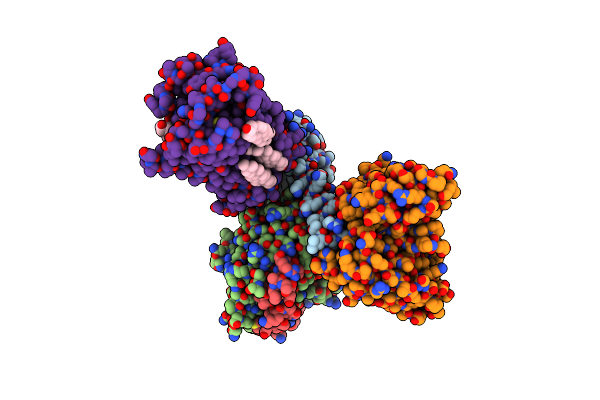 |
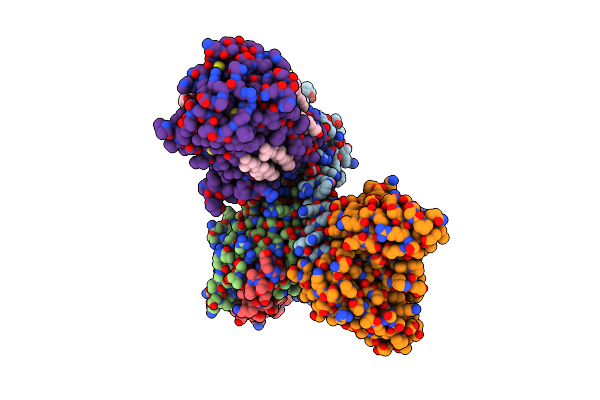
Cryo-Em Structure Of Histamine H3 Receptor In Complex With Proxyfan And Minigo
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: SIGNALING PROTEIN Ligands: A1LY3, CLR |
 |
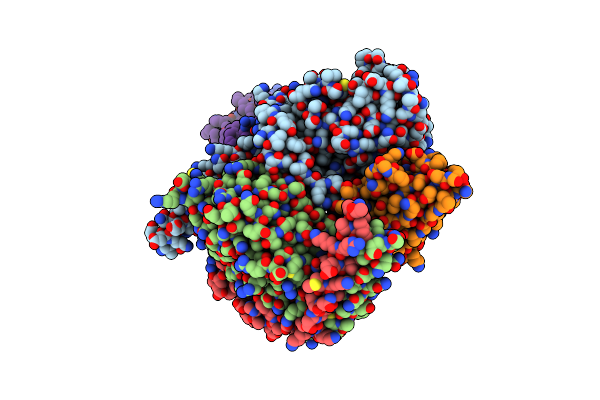
Cryo-Em Structure Of Histamine H1 Receptor In Complex With Histamine And Minigq
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: SIGNALING PROTEIN Ligands: HSM, CLR |
 |
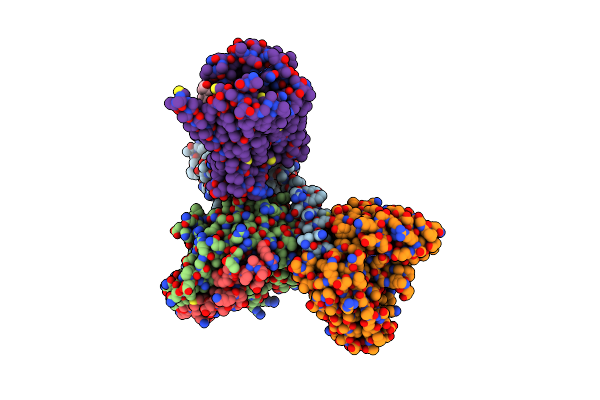
Cryo-Em Structure Of Histamine H2 Receptor In Complex With Histamine And Minigs
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: SIGNALING PROTEIN Ligands: HSM, CLR |
 |
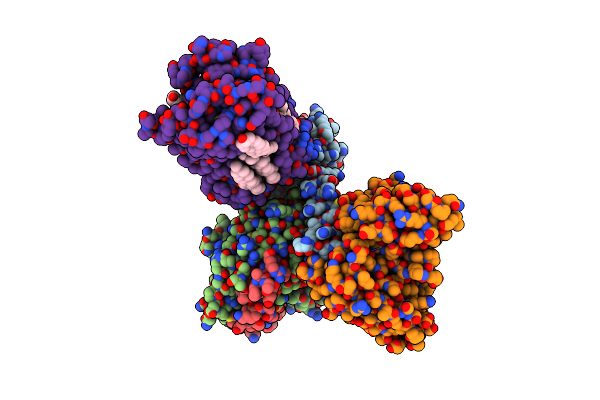
Cryo-Em Structure Of Histamine H2 Receptor In Complex With Histamine And Minigq
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: SIGNALING PROTEIN Ligands: HSM, CLR |
 |
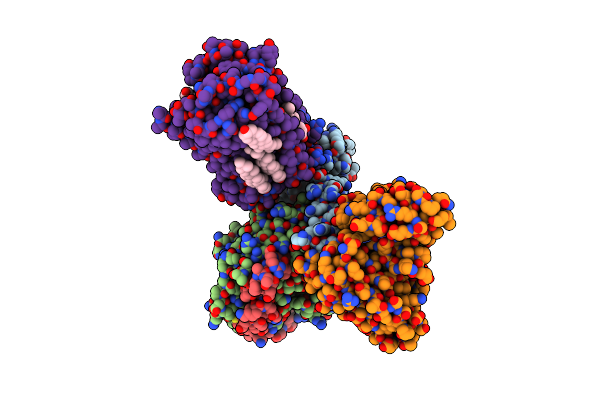
Cryo-Em Structure Of Histamine H3 Receptor In Complex With Histamine And Gi
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: SIGNALING PROTEIN Ligands: HSM, CLR |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: SIGNALING PROTEIN Ligands: ITF, CLR |
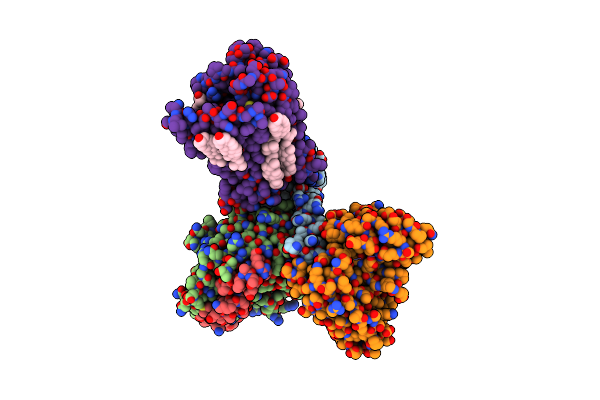 |
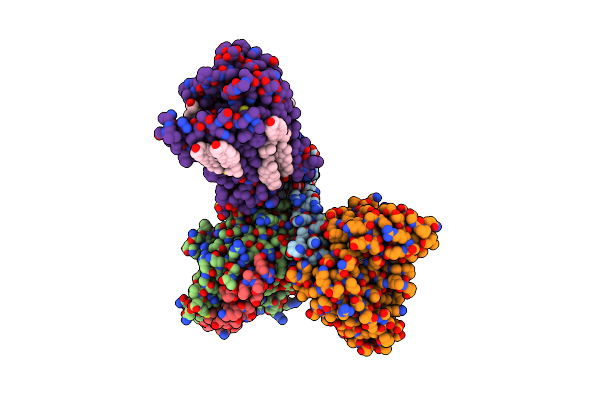
Cryo-Em Structure Of Histamine H4 Receptor In Complex With Histamine And Gi
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: SIGNALING PROTEIN Ligands: HSM, CLR |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: SIGNALING PROTEIN Ligands: A1LY4, CLR |
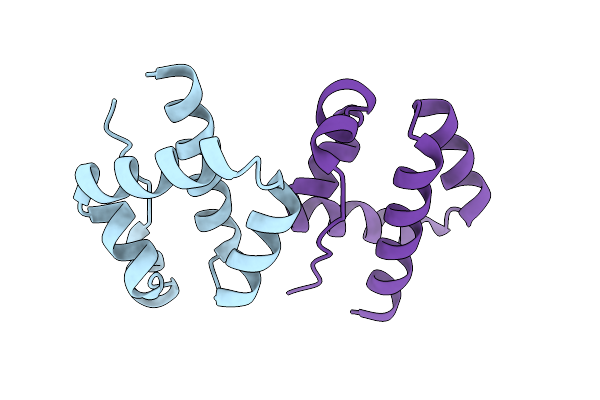 |
Organism: Phytophthora capsici lt1534
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2023-12-06 Classification: IMMUNOSUPPRESSANT |
 |
Organism: Homo sapiens, Lama glama, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Psychromonas sp. b3m02, Synthetic construct, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Lama glama, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of Fenoldopam Bound Dopamine Receptor Drd1-Gs Signaling Complex
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2021-03-03 Classification: CELL CYCLE Ligands: G3C, CLR |
 |
Cryo-Em Structure Of A77636 Bound Dopamine Receptor Drd1-Gs Signaling Complex
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2021-03-03 Classification: CELL CYCLE Ligands: G3O, CLR |
 |
Cryo-Em Structure Of Pw0464 Bound Dopamine Receptor Drd1-Gs Signaling Complex
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2021-03-03 Classification: CELL CYCLE Ligands: CLR, G3U |
 |
Cryo-Em Structure Of Dopamine And Ly3154207 Bound Dopamine Receptor Drd1-Gs Signaling Complex
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2021-03-03 Classification: CELL CYCLE Ligands: CLR, LDP, G4C |
 |
Cryo-Em Structure Of Skf83959 Bound Dopamine Receptor Drd1-Gs Signaling Complex
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2021-03-03 Classification: CELL CYCLE Ligands: GBU |

