Search Count: 40
 |
Crystal Structure Of A Cupin Protein (Tm1459) In Manganese (Mn) Substituted Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: MN |
 |
Crystal Structure Of A Cupin Protein (Tm1459) In Iron (Fe) Substituted Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Crystal Structure Of A Cupin Protein (Tm1459) In Zinc (Zn) Substituted Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of A Cupin Protein (Tm1459, C106V Mutant) In Iron (Fe) Substituted Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: FE |
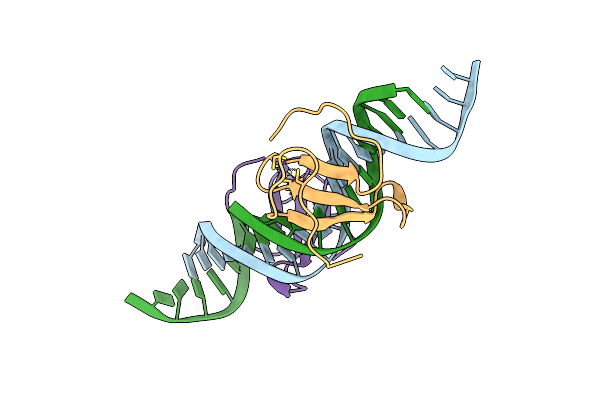 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-03-12 Classification: DNA/DNA BINDING PROTEIN Ligands: ZN |
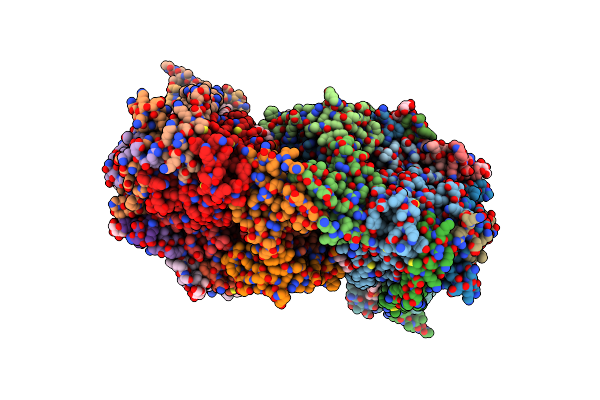 |
Crystal Structure Of Bovine Heart Cytochrome C Oxidase, Apo Structure With Dmso
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, PGV, TGL, CUA, CDL, CHD, PEK, PSC, ZN, DMU |
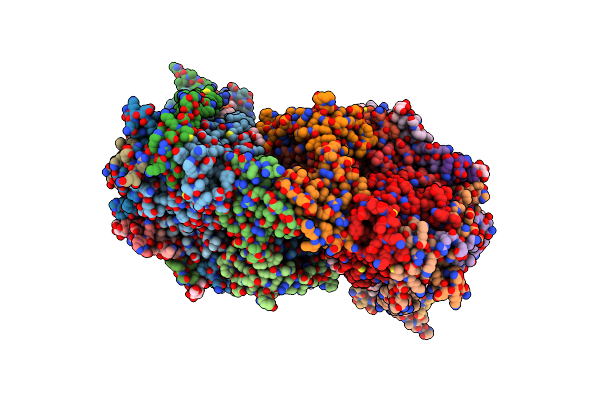 |
Crystal Structure Of Bovine Heart Cytochrome C Oxidase, The Structure Complexed With An Allosteric Inhibitor T113
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, TGL, J6X, CUA, CHD, PSC, PEK, PGV, CDL, DMU, ZN |
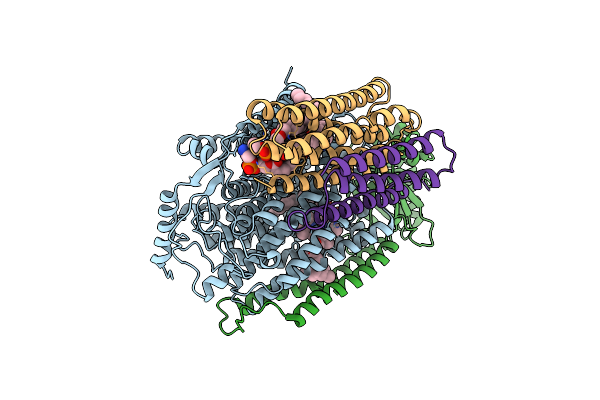 |
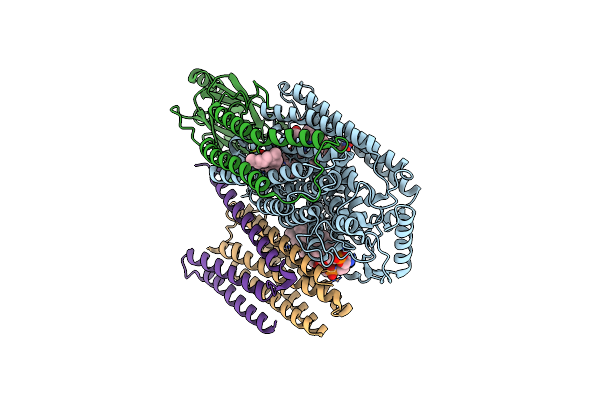
Cryo-Em Structure Of Cytochrome Bo3 From Escherichia Coli, Apo Structure With Dmso
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEO, HEM, CU, PEE, UNX |
 |
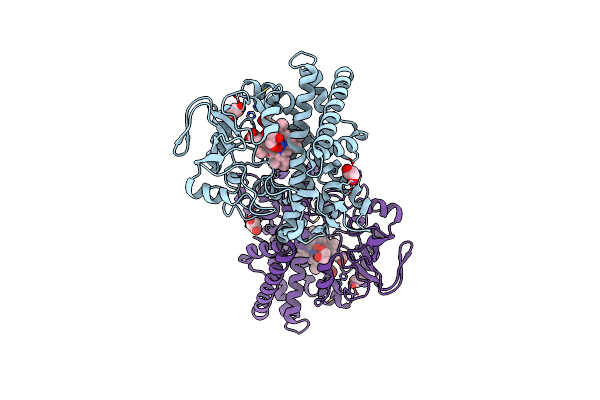
Cryo-Em Structure Of Cytochrome Bo3 From Escherichia Coli, The Structure Complexed With An Allosteric Inhibitor N4
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEO, HEM, CU, PEE, JYR, UNX |
 |
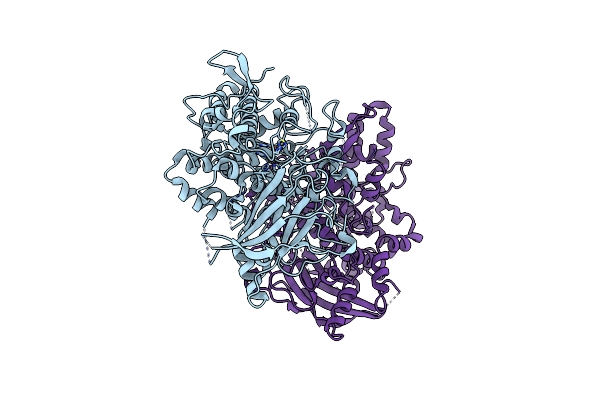
Structure Of Reaction Intermediate Of Cytochrome P450 No Reductase (P450Nor) Determined By Xfel
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-05-19 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
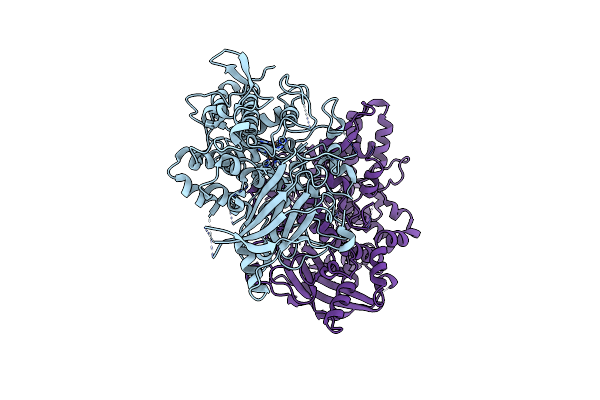
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: NO3 |
 |
Aspergillus Oryzae Active-Tyrosinase Copper-Depleted C92A Mutant Complexed With L-Tyrosine
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: TYR, NO3 |
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU, NO3 |
 |
Aspergillus Oryzae Active-Tyrosinase Copper-Bound C92A Mutant Complexed With L-Tyrosine
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: TYR, DAH, CU, NO3 |
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU, PER |
 |
Radiation Damage In Aspergillus Oryzae Pro-Tyrosinase Oxygen-Bound C92A Mutant
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU, PER |
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU, PER |
 |
Radiation Damage In Aspergillus Oryzae Pro-Tyrosinase Oxygen-Bound C92A/H103F Mutant
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: CU, PER |

