Search Count: 21
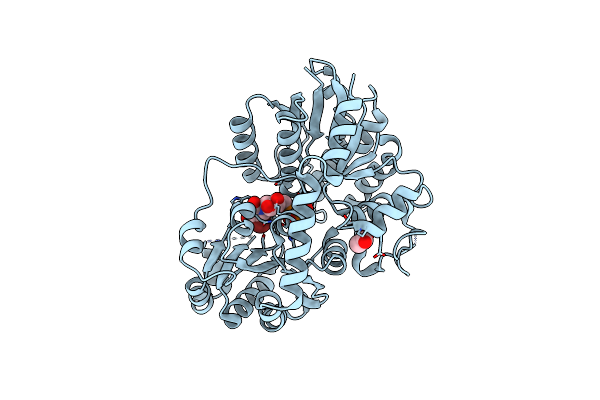 |
Crystal Structure Of A Flavonoid C-Glucosyltrasferase From Fagopyrum Esculentum (Fecgta) Complexed With Brutp
Organism: Fagopyrum esculentum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-09-23 Classification: TRANSFERASE Ligands: ACT, BUP |
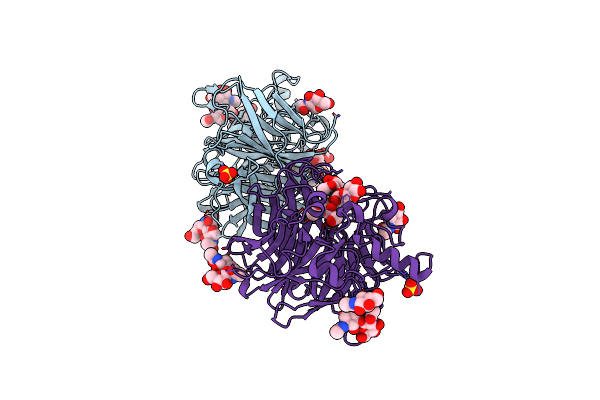 |
Crystal Structure Of Mumps Virus Hemagglutinin-Neuraminidase Bound To The Oligosaccharide Portion Of The Gm2 Ganglioside
Organism: Mumps rubulavirus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-05-22 Classification: VIRAL PROTEIN Ligands: NAG, SO4 |
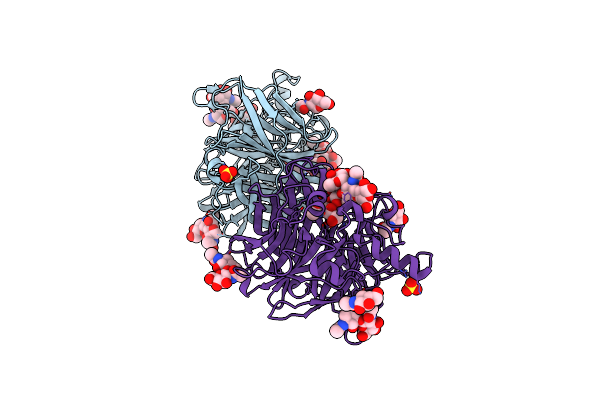 |
Crystal Structure Of Mumps Virus Hemagglutinin-Neuraminidase Bound To Sialyl Lewisx
Organism: Mumps rubulavirus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-05-22 Classification: VIRAL PROTEIN Ligands: NAG, SO4 |
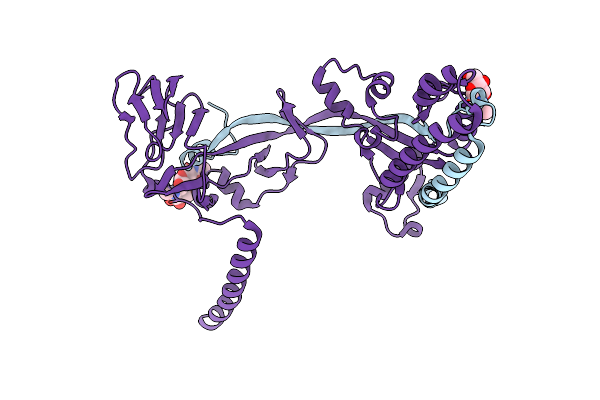 |
Organism: Measles virus (strain ichinose-b95a), Measles virus
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2018-02-21 Classification: VIRAL PROTEIN Ligands: NAG |
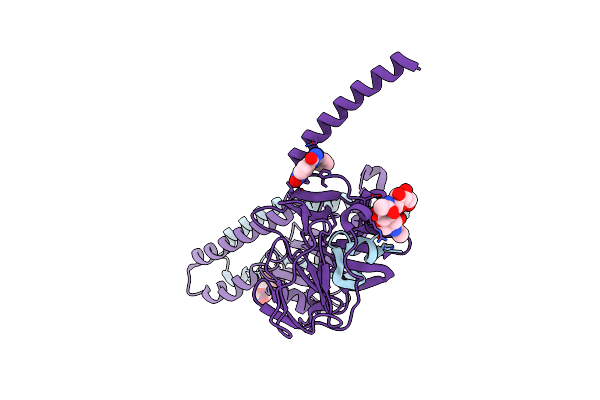 |
Crystal Structure Of The Prefusion Form Of Measles Virus Fusion Protein In Complex With A Fusion Inhibitor Compound (As-48)
Organism: Measles virus (strain ichinose-b95a), Measles virus
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2018-02-21 Classification: VIRAL PROTEIN Ligands: NAG, 95C |
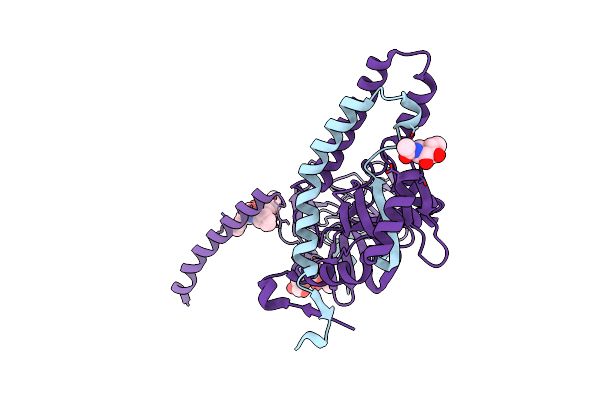 |
Crystal Structure Of The Prefusion Form Of Measles Virus Fusion Protein In Complex With A Fusion Inhibitor Peptide (Fip)
Organism: Measles virus (strain ichinose-b95a), Measles virus, Sendai virus
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2018-02-21 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG |
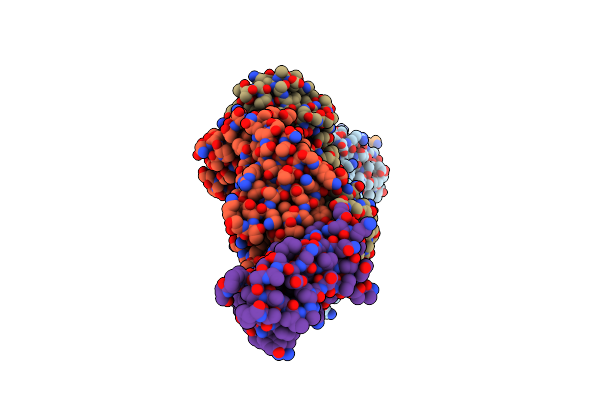 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2017-05-03 Classification: IMMUNE SYSTEM |
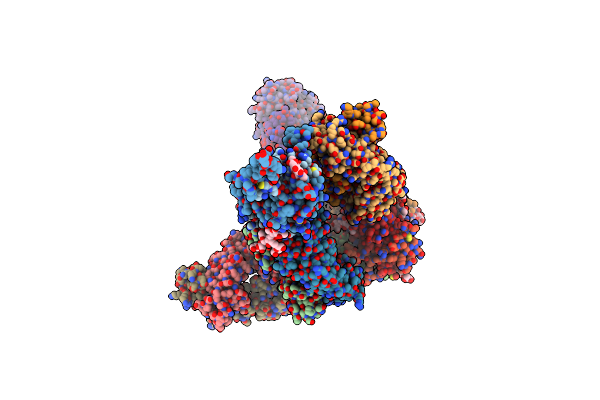 |
Crystal Structure Of Marburg Virus Gp In Complex With The Human Survivor Antibody Mr78
Organism: Lake victoria marburgvirus (strain ravn-87), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2017-03-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, MAN |
 |
Organism: Mumps virus
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2016-09-28 Classification: VIRAL PROTEIN Ligands: NAG, SO4 |
 |
Crystal Structure Of Mumps Virus Hemagglutinin-Neuraminidase Bound To 3-Sialyllactose
Organism: Mumps virus
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2016-09-28 Classification: VIRAL PROTEIN Ligands: NAG, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-11-11 Classification: IMMUNE SYSTEM Ligands: CA |
 |
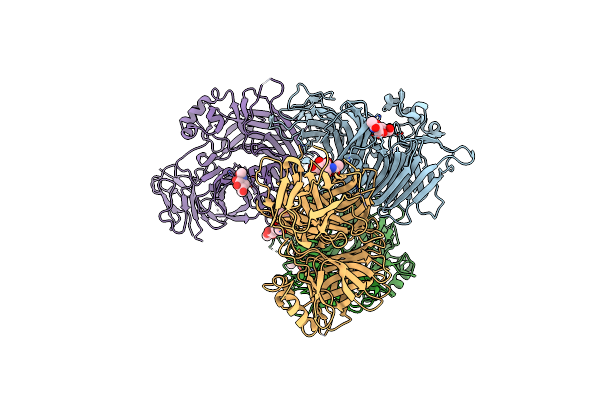
Crystal Structure Of The Measles Virus Hemagglutinin Bound To Its Cellular Receptor Slam (Form I, Mv-H-Slam(N102H/R108Y) Fusion)
Organism: Measles virus, Synthetic construct, Saguinus oedipus
Method: X-RAY DIFFRACTION Resolution:3.55 Å Release Date: 2011-01-12 Classification: viral protein/membrane protein Ligands: NAG |
 |
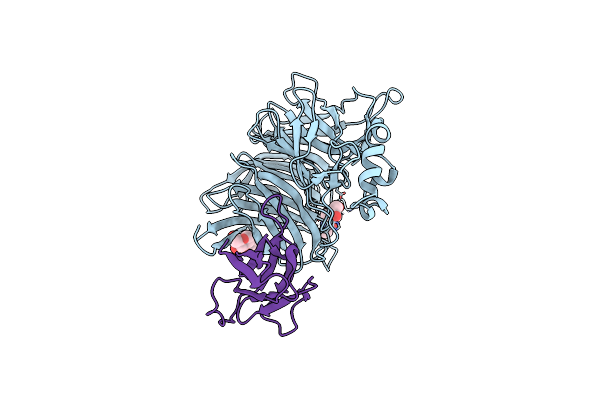
Crystal Structure Of The Measles Virus Hemagglutinin Bound To Its Cellular Receptor Slam (Mv-H(L482R)-Slam(N102H/R108Y) Fusion)
Organism: Measles morbillivirus, Synthetic construct, Saguinus oedipus
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2011-01-12 Classification: Viral Protein/membrane protein Ligands: NAG |
 |
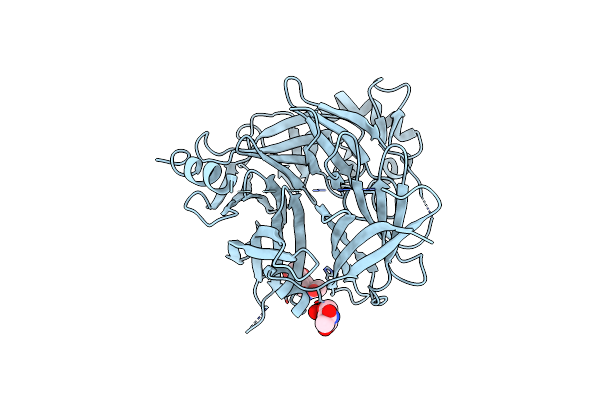
Crystal Structure Of The Measles Virus Hemagglutinin Bound To Its Cellular Receptor Slam (Form I)
Organism: Measles virus, Saguinus oedipus
Method: X-RAY DIFFRACTION Resolution:4.52 Å Release Date: 2011-01-12 Classification: viral protein/membrane protein Ligands: NAG |
 |
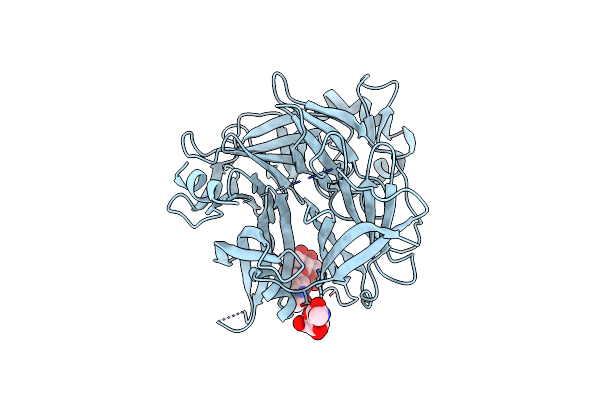
Organism: Measles virus strain edmonston-b
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2007-11-06 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Measles virus strain edmonston-b
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-11-06 Classification: VIRAL PROTEIN Ligands: NAG |
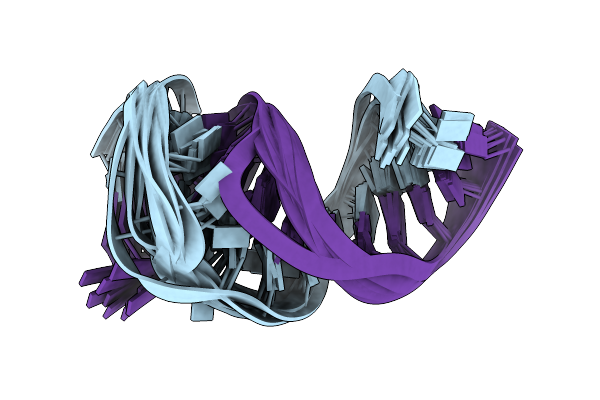 |
Solution Rna Structure Of Stem-Bulge-Stem Region Of The Hiv-1 Dimerization Initiation Site
|
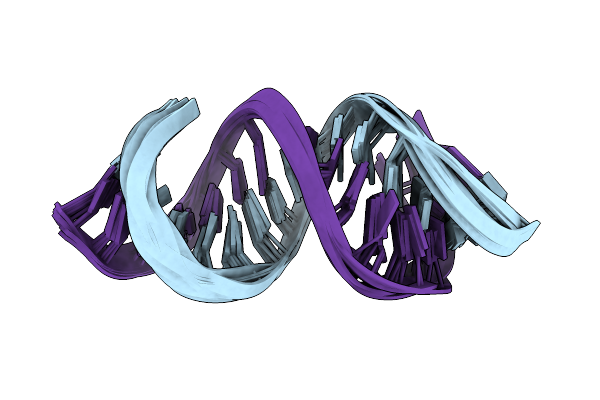 |
Solution Rna Structure Of Loop Region Of The Hiv-1 Dimerization Initiation Site In The Extended-Duplex Dimer
|
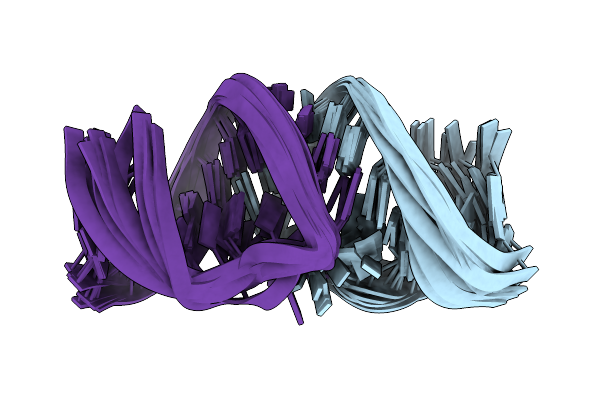 |
Solution Rna Structure Of Loop Region Of The Hiv-1 Dimerization Initiation Site In The Kissing-Loop Dimer
|
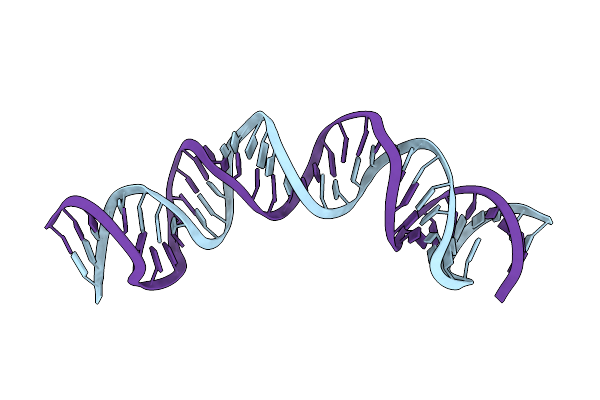 |
Solution Rna Structure Model Of The Hiv-1 Dimerization Initiation Site In The Extended-Duplex Dimer
|

