Search Count: 79
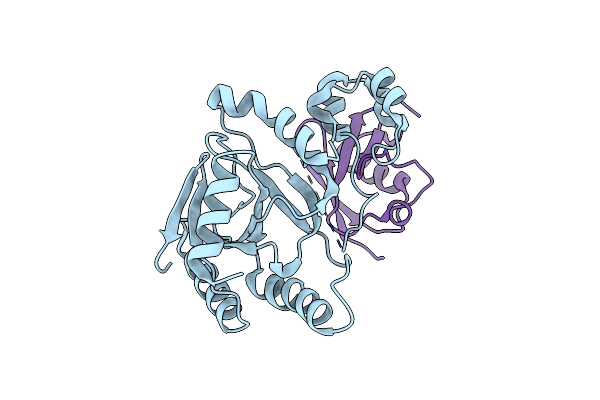 |
Crystal Structure Of A Semet-Labeled Effector From Chromobacterium Violaceum In Complex With Ubiquitin
Organism: Chromobacterium violaceum atcc 12472, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-11-22 Classification: TRANSFERASE |
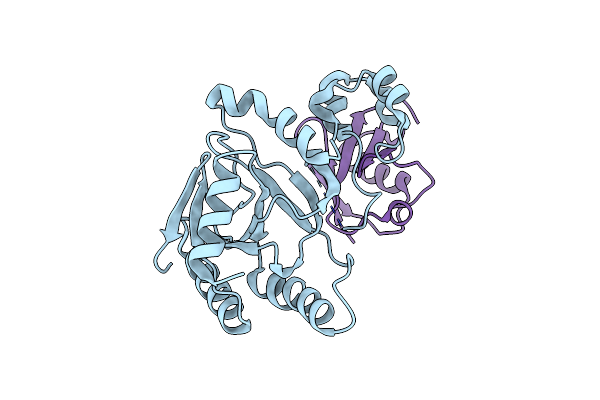 |
Crystal Structure Of An Effector From Chromobacterium Violaceum In Complex With Ubiquitin
Organism: Chromobacterium violaceum atcc 12472, Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-11-22 Classification: TRANSFERASE |
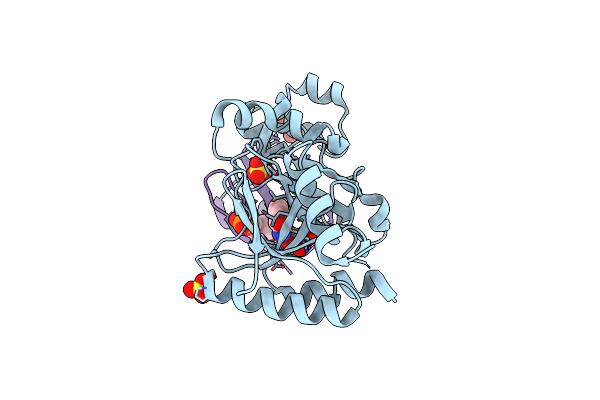 |
Organism: Chromobacterium violaceum atcc 12472, Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: SO4, GOL, NCA, AR6 |
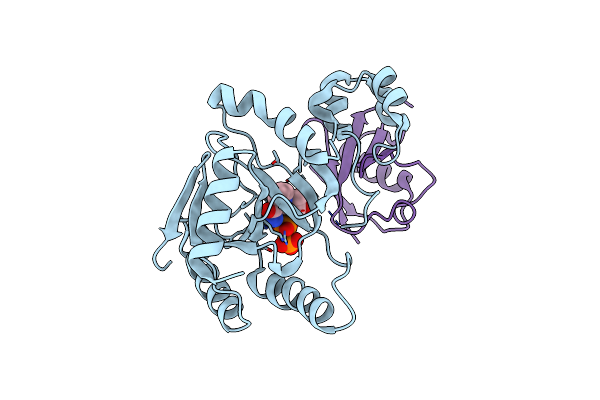 |
Organism: Chromobacterium violaceum atcc 12472, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: NAD |
 |
Crystal Structure Of Dihydroxybenzoate Decarboxylase Mutant A63S From Aspergillus Oryzae In Complex With Catechol
Organism: Aspergillus oryzae rib40
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-01-18 Classification: HYDROLASE Ligands: MG, CAQ |
 |
Crystal Structure Of Sugar Binding Protein Cbpa From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Sugar Binding Protein Cbpa Complexed Wtih Glucose From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN Ligands: BGC |
 |
Crystal Structure Of Sugar Binding Protein Cbpb From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Sugar Binding Protein Cbpb Complexed Wtih Cellobiose From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Sugar Binding Protein Cbpb Complexed Wtih Cellotriose From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Sugar Binding Protein Cbpb Complexed Wtih Cellotetraose From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Sugar Binding Protein Cbpb Complexed Wtih Cellopentaose From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Sugar Binding Protein Cbpb Complexed Wtih Laminaribiose From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Sugar Binding Protein Cbpc From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Sugar Binding Protein Cbpd From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of Substrate Binding Protein Lbp Complexed Wtih Guanosine From Clostridium Thermocellum
Organism: Acetivibrio thermocellus
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-09-14 Classification: SUGAR BINDING PROTEIN Ligands: GMP, ZN |
 |
Organism: Rabies virus nishigahara rceh
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-04-20 Classification: VIRAL PROTEIN Ligands: SO4 |
 |
Organism: Rabies virus nishigahara rceh
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-04-20 Classification: VIRAL PROTEIN Ligands: EDO, SO4, PO4 |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-03-16 Classification: NUCLEAR PROTEIN Ligands: GOL |

