Search Count: 17
 |
Crystal Structure Of Amp-Pnp Bound Mutant A3B3 Complex From Enterococcus Hirae V-Atpase
Organism: Enterococcus hirae (strain atcc 9790 / dsm 20160 / jcm 8729 / lmg 6399 / nbrc 3181 / ncimb 6459 / ncdo 1258)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: ANP, MG, GOL, MES |
 |
Organism: Enterococcus hirae (strain atcc 9790 / dsm 20160 / jcm 8729 / lmg 6399 / nbrc 3181 / ncimb 6459 / ncdo 1258)
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Enterococcus hirae atcc 9790
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2016-11-02 Classification: HYDROLASE Ligands: MG, ADP, GOL |
 |
Organism: Enterococcus hirae atcc 9790
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2016-11-02 Classification: HYDROLASE Ligands: MG, SO4, ADP, GOL |
 |
Organism: Enterococcus hirae atcc 9790
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2016-11-02 Classification: HYDROLASE Ligands: MG, GOL, PO4, B3P |
 |
Crystal Structure Of Nucleotide-Free A3B3 Complex From Enterococcus Hirae V-Atpase [Ea3B3]
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-01-16 Classification: HYDROLASE |
 |
Crystal Structure Of Amp-Pnp Bound A3B3 Complex From Enterococcus Hirae V-Atpase [Ba3B3]
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: ANP, MG |
 |
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: GOL, CL, B3P |
 |
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2013-01-16 Classification: HYDROLASE |
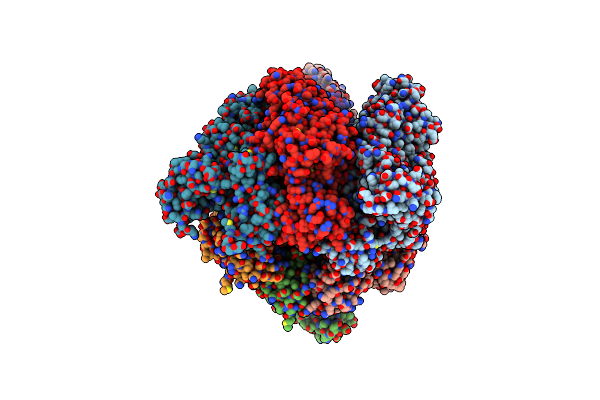 |
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: ANP, MG |
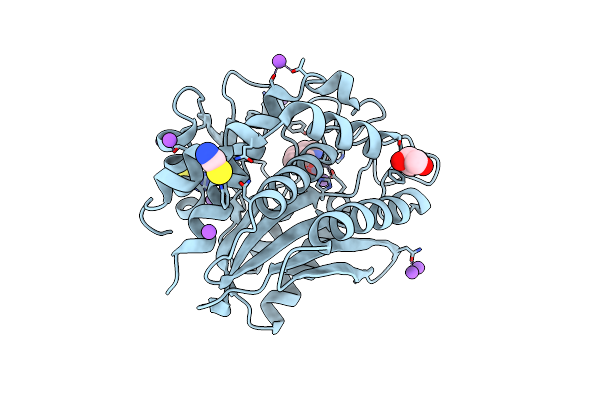 |
Crystal Structure Of Aeromonas Proteolytica Aminopeptidase Complexed With 8-Quinolinol
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2012-05-02 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, HQY, NA, CL, SCN, GOL |
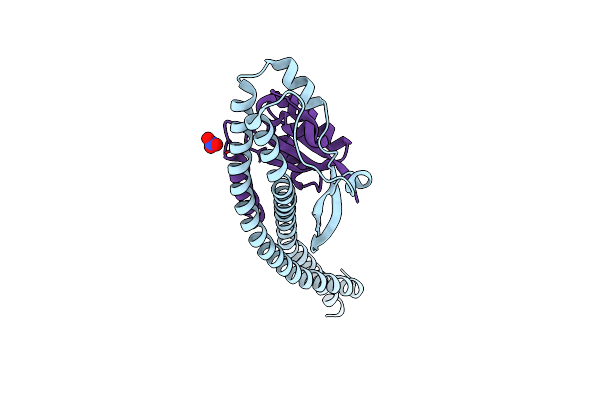 |
Crystal Structure Of The Central Axis (Ntpd-Ntpg) In The Catalytic Portion Of Enterococcus Hirae V-Type Sodium Atpase
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-10-05 Classification: HYDROLASE Ligands: NO3 |
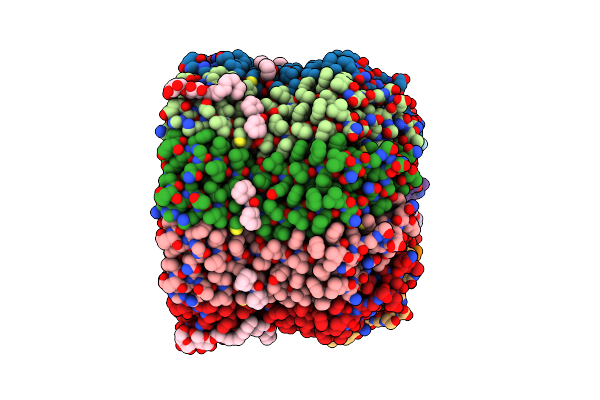 |
Structure Of The Na+ Unbound Rotor Ring Modified With N,N F-Dicyclohexylcarbodiimide Of The Na+-Transporting V-Atpase
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2011-08-17 Classification: HYDROLASE Ligands: DCW, UMQ |
 |
Crystal Structure Of Rotor Ring With Dccd Of The V- Atpase From Enterococcus Hirae
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-12-05 Classification: HYDROLASE Ligands: DCW, NA, LHG, UMQ |
 |
Crystal Structure Of Lithium Bound Rotor Ring Of The V-Atpase From Enterococcus Hirae
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2006-06-27 Classification: HYDROLASE Ligands: LI, LHG, UMQ |
 |
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-04-05 Classification: HYDROLASE Ligands: LHG, NA, UMQ |
 |
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2003-07-29 Classification: LYASE Ligands: MG, SO4, GOL |

