Search Count: 34
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: NUCLEAR PROTEIN Ligands: A1MAM, EDO, ACT |
 |
Organism: Vibrio alginolyticus
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2021-07-07 Classification: MOTOR PROTEIN |
 |
Crystal Structure Of Flim Middle Domain (46-231) With R49P Substitution From Vibro Alginolyticus
Organism: Vibrio alginolyticus
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2021-07-07 Classification: MOTOR PROTEIN |
 |
Organism: Eucommia ulmoides
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-02-17 Classification: PLANT PROTEIN |
 |
Organism: Eucommia ulmoides
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2021-02-17 Classification: PLANT PROTEIN |
 |
Organism: Eucommia ulmoides
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2021-02-17 Classification: PLANT PROTEIN |
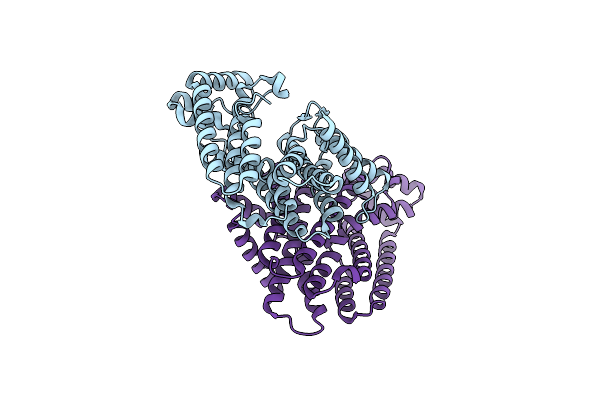 |
Organism: Eucommia ulmoides
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-02-17 Classification: PLANT PROTEIN |
 |
Organism: Talaromyces funiculosus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: NAG, GOL, CIT, PEG, PGE |
 |
The Complex Structure Of Endo-Beta-1,2-Glucanase From Talaromyces Funiculosus With Sophorose
Organism: Talaromyces funiculosus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: NAG, BGC, PEG |
 |
The Complex Structure Of Endo-Beta-1,2-Glucanase Mutant (E262Q) From Talaromyces Funiculosus With Beta-1,2-Glucan
Organism: Talaromyces funiculosus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: NAG, PEG |
 |
Crystal Structure Of The Fab Fragment Of B5209B, A Murine Monoclonal Antibody Specific For The Fifth Immunoglobulin Domain (Ig5) Of Human Robo1
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-01-30 Classification: IMMUNE SYSTEM Ligands: GOL, SO4 |
 |
Crystal Structure Of The Fifth Immunoglobulin Domain (Ig5) Of Human Robo1 In Complex With The Fab Fragment Of Murine Monoclonal Antibody B5209B
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-30 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of The Fifth Immunoglobulin Domain (Ig5) Of Human Robo1 In Complex With The Scfv Fragment Of Murine Monoclonal Antibody B5209B
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-01-30 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Crystal Structure Of The Fifth Immunoglobulin Domain (Ig5) Of Human Robo1 In Complex With The Mutant Scfv Fragment (P103A) Of Murine Monoclonal Antibody B5209B
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2019-01-30 Classification: IMMUNE SYSTEM Ligands: SO4 |
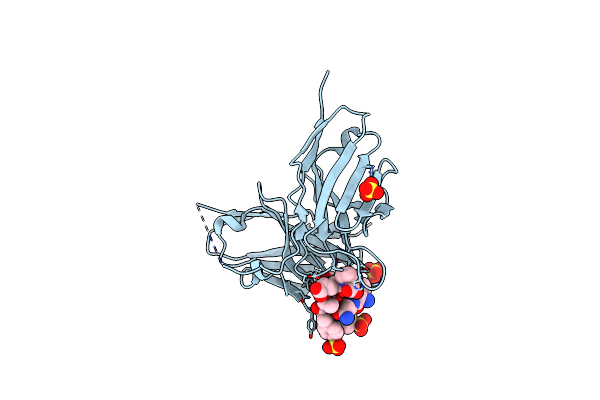 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2018-07-18 Classification: IMMUNE SYSTEM Ligands: SO4, CL |
 |
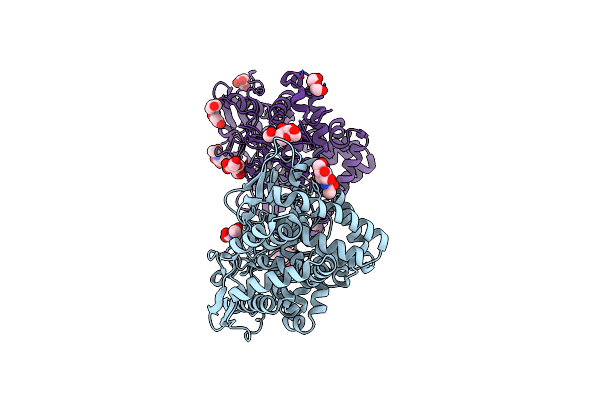
Structure Of The Ndi1 Protein From Saccharomyces Cerevisiae In Complex With The Competitive Inhibitor, Stigmatellin.
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-02-14 Classification: OXIDOREDUCTASE Ligands: FAD, MG, PE4, P6G, GOL, PG4, PEG, PGE, MES, SMA |
 |
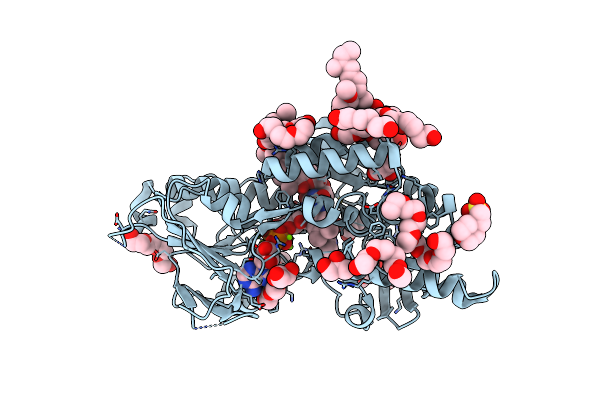
Structure Of The Ndi1 Protein From Saccharomyces Cerevisiae In Complex With Myxothiazol.
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2018-02-14 Classification: OXIDOREDUCTASE Ligands: FAD, MG, MES, MYX |
 |
Structure Of The Ndi1 Protein From Saccharomyces Cerevisiae In Complex With Ac0-12.
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-02-14 Classification: OXIDOREDUCTASE Ligands: FAD, MG, 8WF, MES |
 |
Organism: Chitinophaga pinensis (strain atcc 43595 / dsm 2588 / ncib 11800 / uqm 2034)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-03-15 Classification: HYDROLASE Ligands: IOD |
 |
Endo-Beta-1,2-Glucanase From Chitinophaga Pinensis - Sophorotriose And Glucose Complex
Organism: Chitinophaga pinensis (strain atcc 43595 / dsm 2588 / ncib 11800 / uqm 2034)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-03-15 Classification: HYDROLASE Ligands: PO4, K, BGC, PG4, PEG, CL, 1PE |

