Search Count: 178
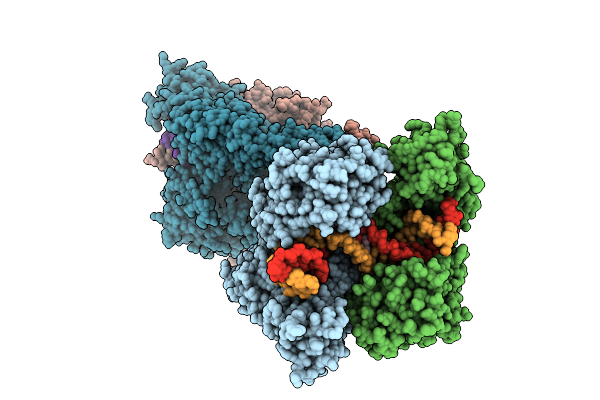 |
Crystal Structure Of Topoisomerase Iv From Klebsiella Pneumoniae In Complex With Dna And Bwc0977, A Dual-Targeting Broad-Spectrum Novel Bacterial Topoisomerase Inhibitor.
Organism: Klebsiella pneumoniae subsp. pneumoniae mgh 78578, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2025-11-12 Classification: ISOMERASE/DNA/ISOMERASE INHIBITOR Ligands: A1L5V |
 |
Crystal Structure Of Aminoglycoside Efflux Transporter Mexy From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN Ligands: SO4 |
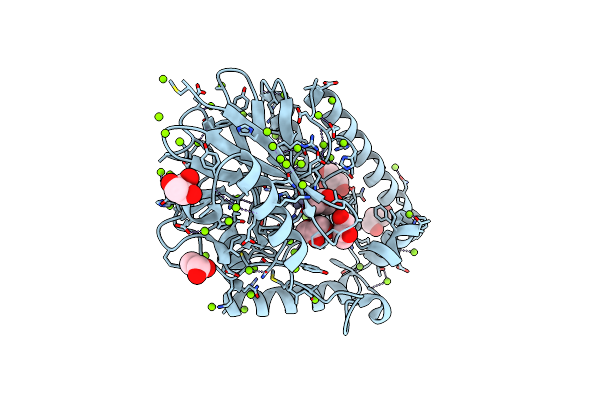 |
Organism: Bacillus sp. (in: firmicutes)
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-06-18 Classification: HYDROLASE Ligands: GOL, MG, ZN |
 |
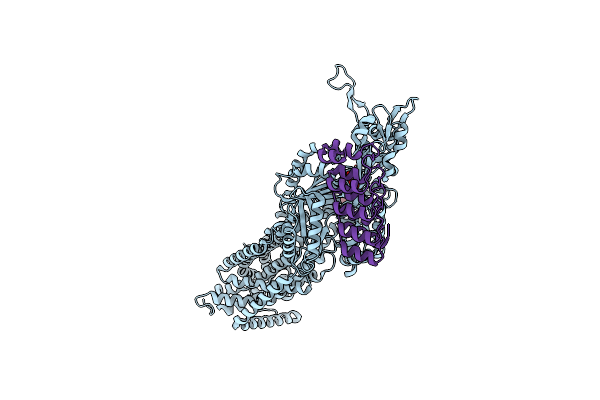
Crystal Structure Of Multidrug Efflux Transporter Oqxb From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-06-18 Classification: MEMBRANE PROTEIN Ligands: LMT, GOL |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN Ligands: MIY |
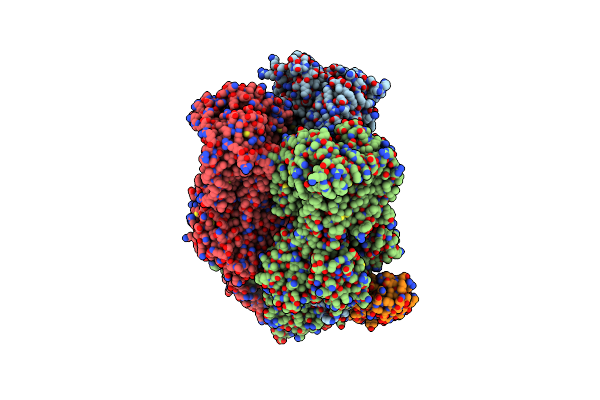 |
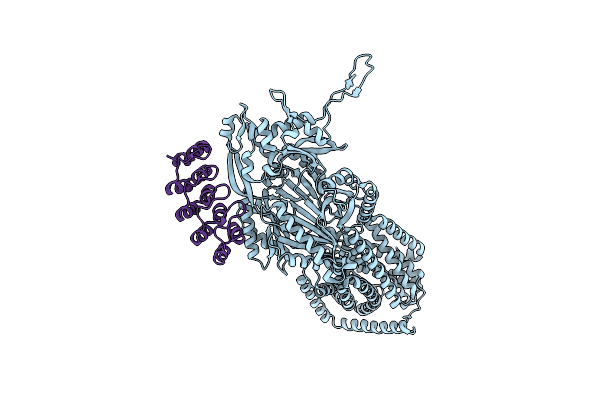
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN |
 |
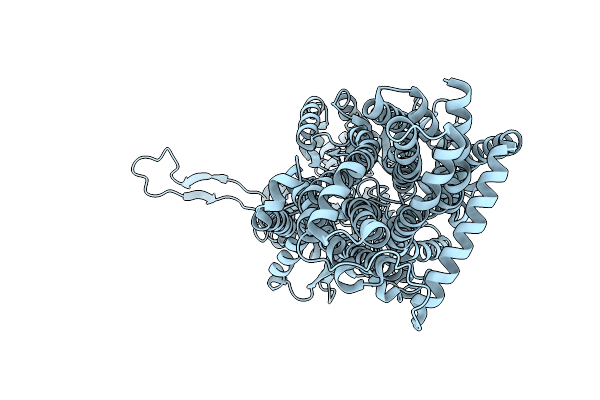
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.55 Å Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN |
 |
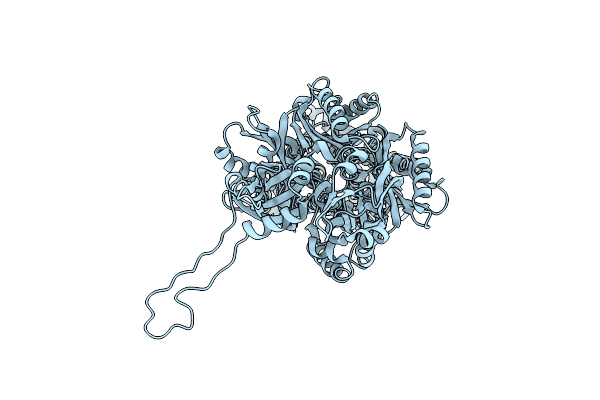
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
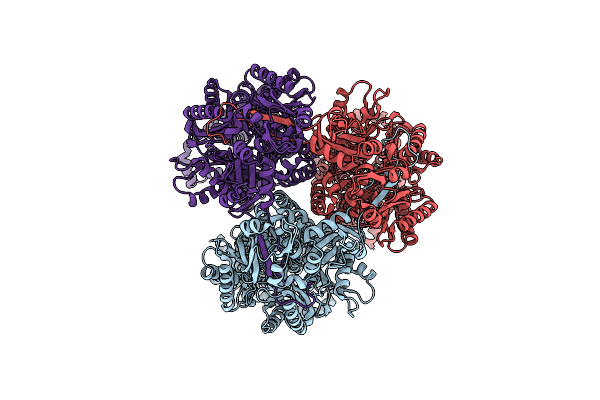
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
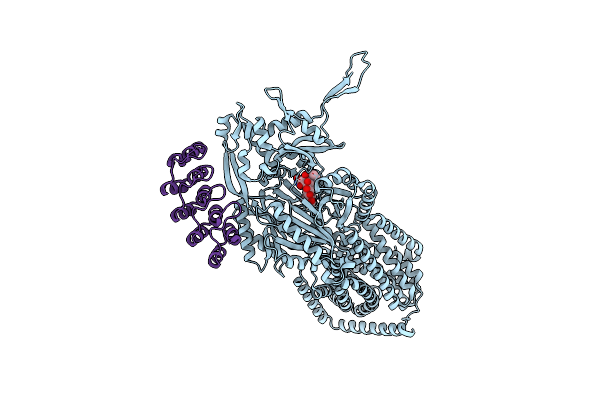
Single Particle Cryo-Em Structure Of The Multidrug Efflux Pump Oqxb From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
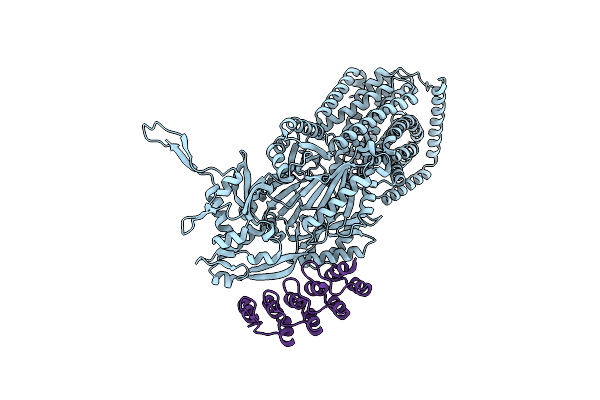
Organism: Synthetic construct, Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN Ligands: MIY |
 |
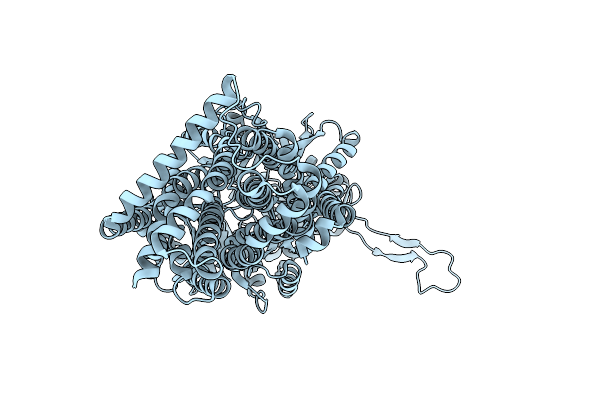
Organism: Synthetic construct, Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
De Novo Designed Cell-Penetrating Peptide Self-Assembly Featuring Distinctive Tertiary Structure
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-09-11 Classification: DE NOVO PROTEIN Ligands: GOL |
 |
Organism: Escherichia phage mu
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-02-28 Classification: VIRAL PROTEIN |
 |
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: ATP, MG |
 |
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: ATP, MG |
 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-09-20 Classification: PROTEIN BINDING |
 |
Crystal Structure Of The Multidrug Efflux Transporter Bpeb From Burkholderia Pseudomallei
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2023-07-19 Classification: MEMBRANE PROTEIN Ligands: UMQ, PG4 |
 |
Crystal Structure Of The Multidrug Effulx Transporter Bpef From Burkholderia Pseudomallei.
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-07-19 Classification: MEMBRANE PROTEIN Ligands: LMT |

