Search Count: 38
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:0.89 Å Release Date: 2024-05-15 Classification: PLANT PROTEIN Ligands: PEG |
 |
Crystal Structure Of L182E D-Succinylase (Dsa) From Cupriavidus Sp. P4-10-C
Organism: Cupriavidus sp. p4-10-c
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-01-26 Classification: HYDROLASE Ligands: CAD, GOL, SIN, CO3, CA, CL |
 |
Organism: Cupriavidus sp. p4-10-c
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-01-26 Classification: HYDROLASE Ligands: GOL, CO3, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2018-11-21 Classification: signaling protein/antagonist Ligands: CF7 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2018-08-01 Classification: signaling protein/agonist Ligands: CFJ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2018-03-21 Classification: transcription/agonist Ligands: MPD, E3S |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-03-21 Classification: transcription/agonist Ligands: MPD, E3V |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2018-01-03 Classification: SIGNALING PROTEIN Ligands: CFG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-07-18 Classification: BLOOD CLOTTING Ligands: NAG |
 |
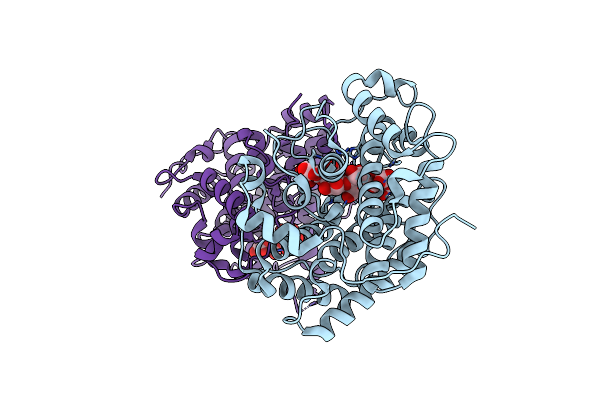
Organism: Sphingomonas
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-06-27 Classification: LYASE |
 |
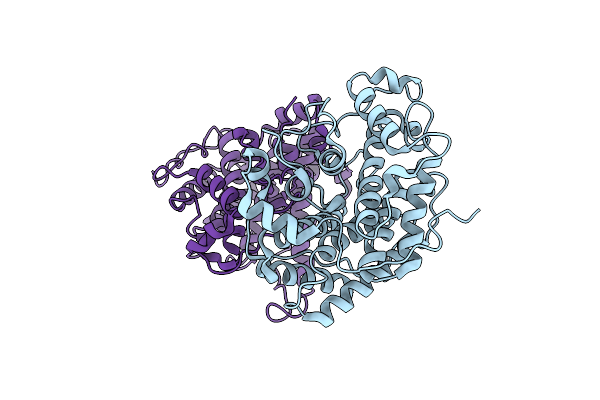
Organism: Sphingomonas
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2012-06-27 Classification: LYASE |
 |
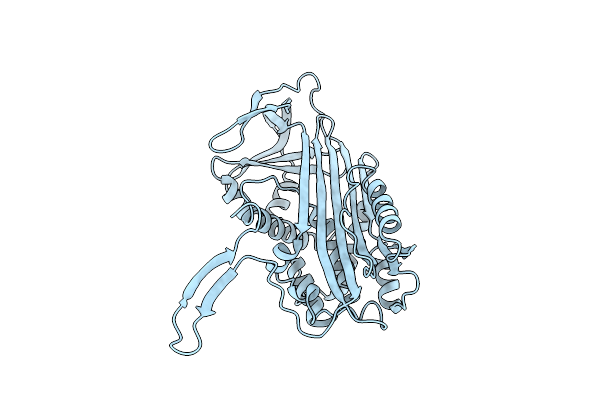
Organism: Sphingomonas
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-04-11 Classification: LYASE |
 |
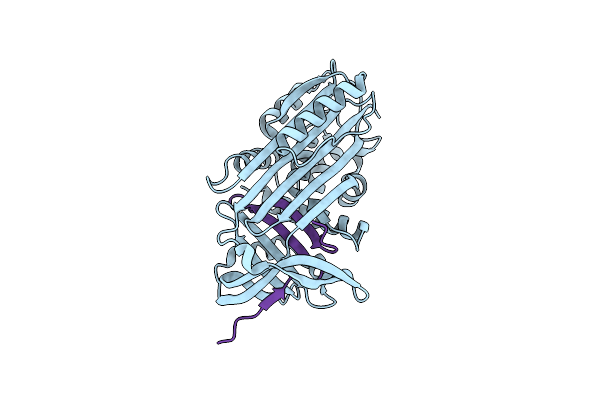
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2011-08-17 Classification: HYDROLASE inhibitor |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-07-28 Classification: HYDROLASE INHIBITOR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-07-28 Classification: HYDROLASE INHIBITOR |
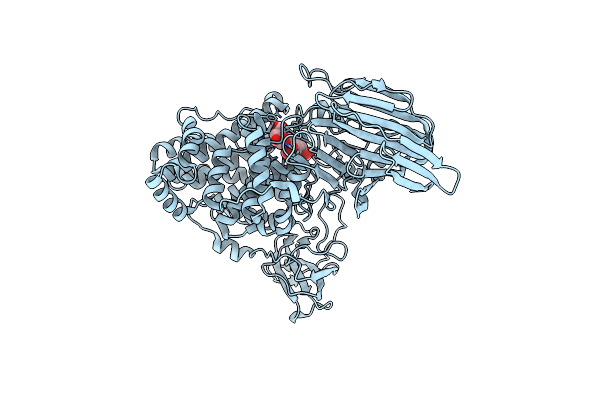 |
Crystal Structure Of Exotype Alginate Lyase Atu3025 H531A Complexed With Alginate Trisaccharide
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2010-04-28 Classification: LYASE |
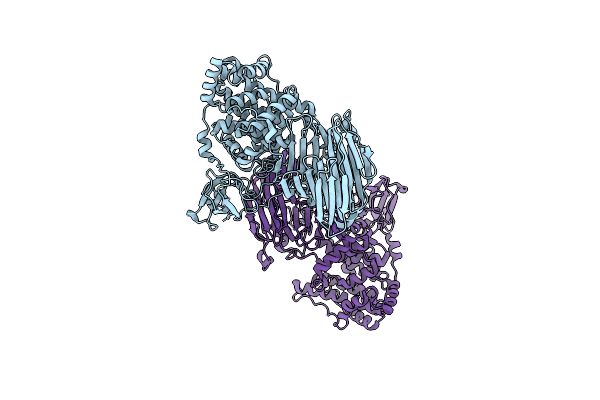 |
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2010-03-31 Classification: LYASE Ligands: CL |
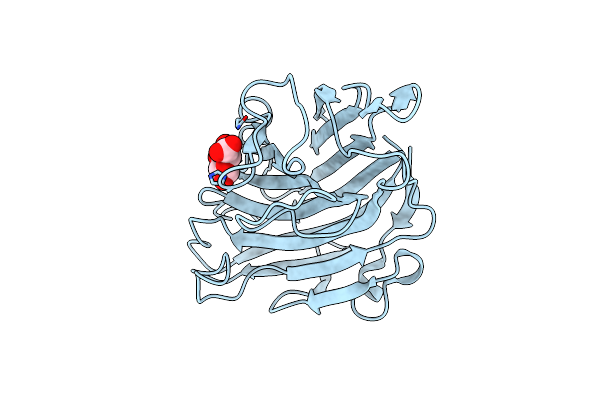 |
Crystal Structure Of D-Glucuronic Acid-Bound Alginate Lyase Val-1 From Chlorella Virus
Organism: Chlorella virus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2009-10-20 Classification: LYASE Ligands: BDP |
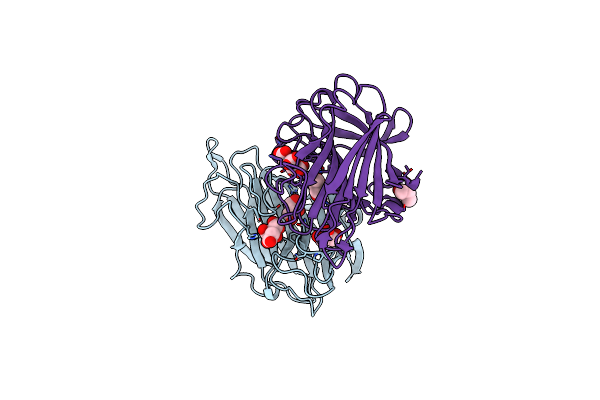 |
Organism: Chlorella virus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2009-10-20 Classification: LYASE Ligands: FLC, GOL |
 |
Crystal Structure Of Chlorella Virus Val-1 Soaked In 200Mm D-Glucuronic Acid, 10% Peg-3350, And 200Mm Glycine-Naoh (Ph 10.0)
Organism: Chlorella virus
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2009-10-20 Classification: LYASE Ligands: BDP |

