Search Count: 16
 |
Organism: Schizosaccharomyces pombe, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2022-11-02 Classification: PROTEIN TRANSPORT |
 |
Crystal Structure Of Fusion Protein Of Human Tp53Inp2 Lir And Human Gabarap
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-11-02 Classification: PROTEIN TRANSPORT Ligands: IPA, PO4 |
 |
Crystal Structure Of Gabarap Complexed With The Tex264 Lir Phosphorylated At Ser271 And Ser272
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-03-30 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-03-30 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2020-07-08 Classification: MEMBRANE PROTEIN Ligands: ALE, EDO |
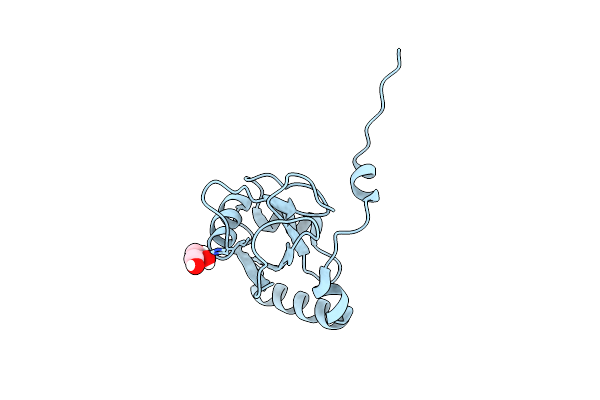 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-07-08 Classification: MEMBRANE PROTEIN Ligands: GOL |
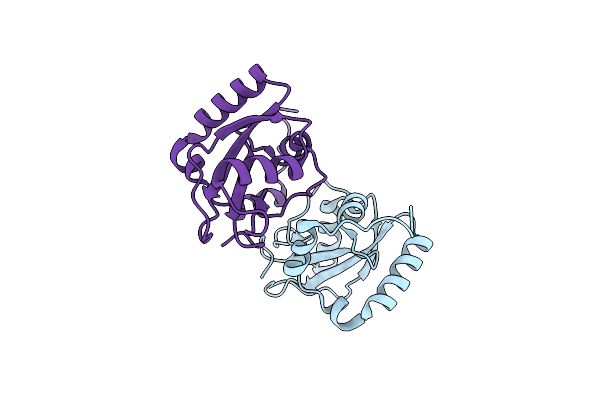 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-08 Classification: MEMBRANE PROTEIN |
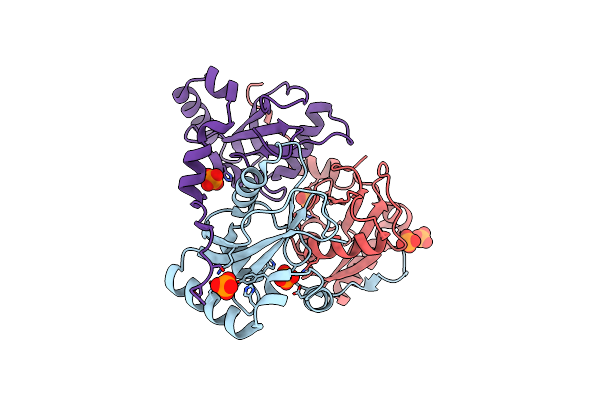 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-07-08 Classification: MEMBRANE PROTEIN Ligands: PO4 |
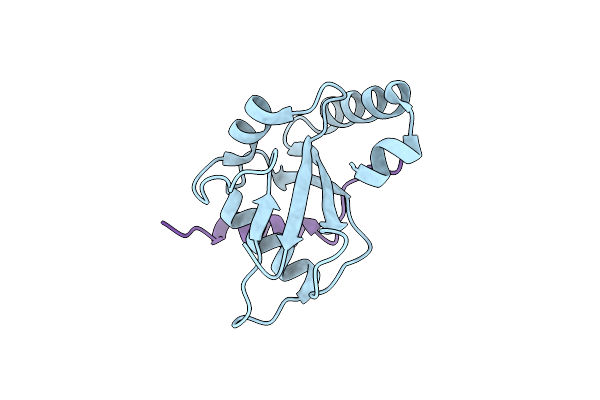 |
Crystal Structure Of Fission Yeast Atg8 Complexed With The Helical Aim Of Hfl1.
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-12-12 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of Budding Yeast Atg8 Complexed With The Helical Aim Of Hfl1.
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae (strain yjm789)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-12-12 Classification: MEMBRANE PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2016-06-29 Classification: PROTEIN TRANSPORT |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2016-06-29 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-06-29 Classification: HYDROLASE Ligands: ZN, CAC |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-06-29 Classification: HYDROLASE |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-04-03 Classification: OXIDOREDUCTASE Ligands: CU, MG, NA, HEA, PGV, CUA, TGL, PSC, CHD, DMU, UNX, PEK, CDL, ZN |