Search Count: 28
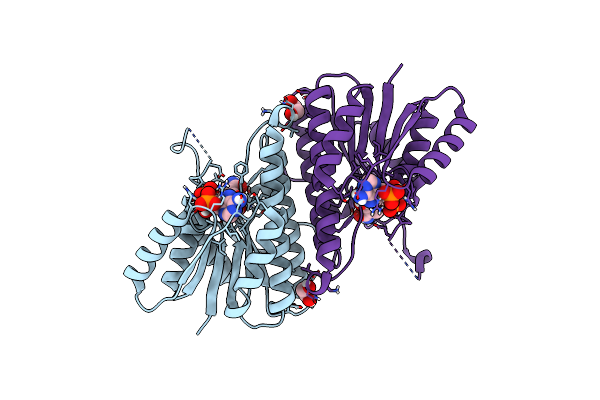 |
Organism: Clostridium acetobutylicum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: NAP, EDO |
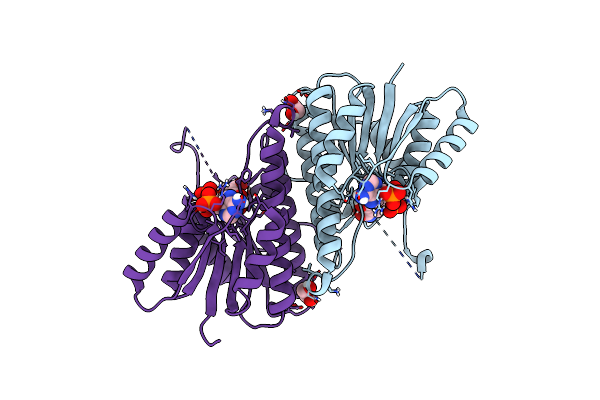 |
Clostridium Acetobutylicum Alcohol Dehydrogenase Bound To Nadp+, Disordered Nicotinamide
Organism: Clostridium acetobutylicum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: NAP, EDO |
 |
Crystal Structure Of The Ethidium Bound Ramr Determined With Xtalab Synergy
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. 14028s
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-10-09 Classification: DNA BINDING PROTEIN Ligands: ET, SO4 |
 |
Crystal Structure Of The Cholic Acid Bound Ramr Determined With Xtalab Synergy
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. 14028s
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-10-09 Classification: DNA BINDING PROTEIN Ligands: CHD, SO4 |
 |
Crystal Structure Of The Gefitinib Intermediate 1 Bound Ramr Determined With Xtalab Synergy
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. 14028s
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-10-09 Classification: DNA BINDING PROTEIN Ligands: XZ1, SO4 |
 |
Crystal Structure Of Ha33 From Clostridium Botulinum Serotype C Strain Yoichi
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-06-15 Classification: SUGAR BINDING PROTEIN Ligands: PGE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:0.96 Å Release Date: 2014-11-12 Classification: HYDROLASE Ligands: NO3, NA, EDO |
 |
Crystal Structure Analysis Of Cyanidioschyzon Melorae Ferredoxin D58N Mutant
Organism: Cyanidioschyzon merolae
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2013-08-07 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
Crystal Structure Of Nontoxic Nonhemagglutinin Subcomponent (Ntnha) From Clostridium Botulinum Serotype D Strain 4947
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2012-09-19 Classification: TOXIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2012-09-05 Classification: TRANSFERASE |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2008-08-19 Classification: LIGASE |
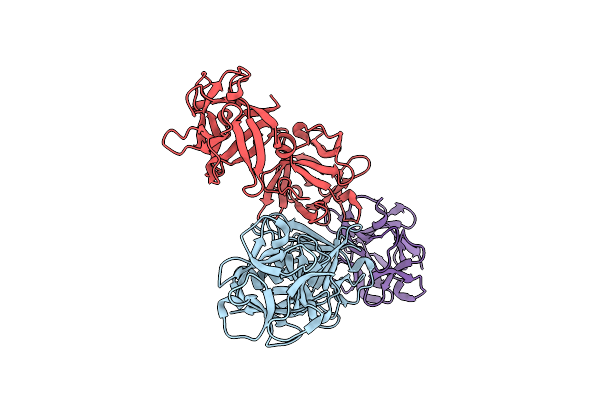 |
Crystal Structure Of Hemagglutinin Subcomponent Complex (Ha-33/Ha-17) From Clostridium Botulinum Serotype D Strain 4947
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-06-19 Classification: TOXIN |
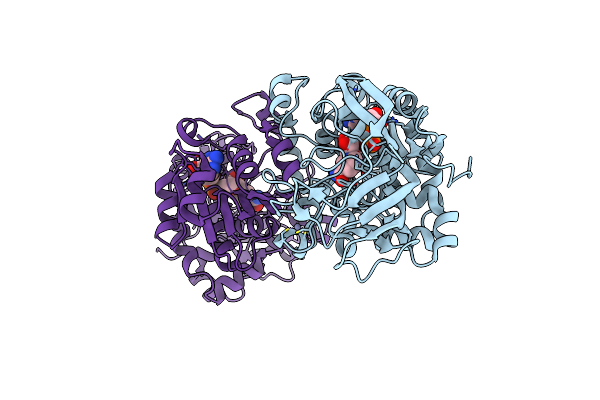 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2007-05-29 Classification: OXIDOREDUCTASE Ligands: ZN, NAD |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2006-07-11 Classification: UNKNOWN FUNCTION Ligands: IOD |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-07-11 Classification: UNKNOWN FUNCTION Ligands: IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-07-11 Classification: HYDROLASE Ligands: CA, IOD |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-07-11 Classification: HYDROLASE Ligands: IOD |
 |
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2006-07-11 Classification: HYDROLASE |
 |
Organism: Hypocrea jecorina
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-07-11 Classification: HYDROLASE Ligands: IOD |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-10-16 Classification: OXIDOREDUCTASE |

