Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
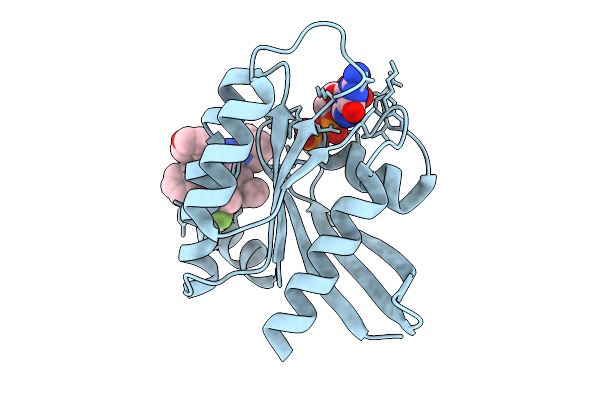 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE Ligands: GDP, MG, A1L65 |
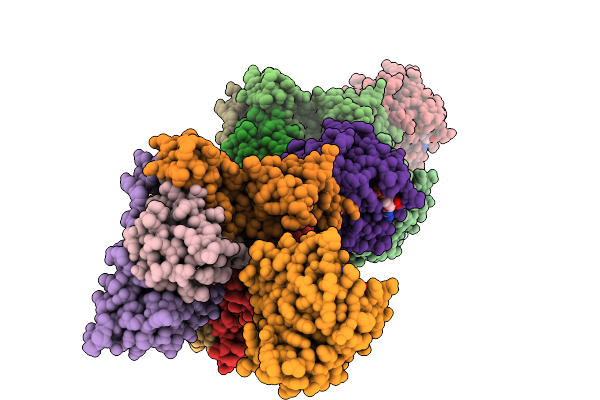 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE Ligands: A1L66, GDP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-01-17 Classification: HYDROLASE/INHIBITOR Ligands: GDP, MG, Y9D |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2022-08-10 Classification: ONCOPROTEIN/INHIBITOR Ligands: GDP, MG, IQC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-08-10 Classification: ONCOPROTEIN/inhibitor Ligands: GDP, MG, SO4, IQN |
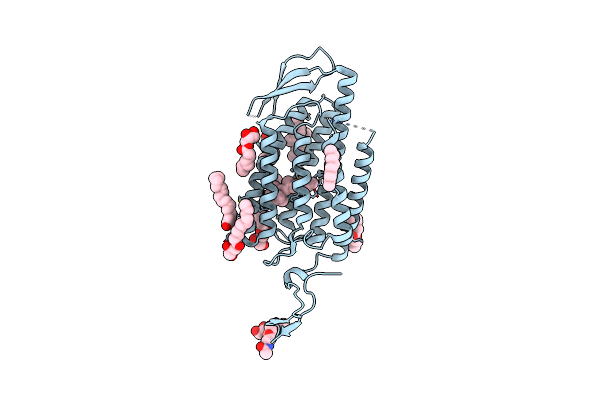 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: Dark State Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 4 Ms Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 1 Microsecond Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 50 Microsecond Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
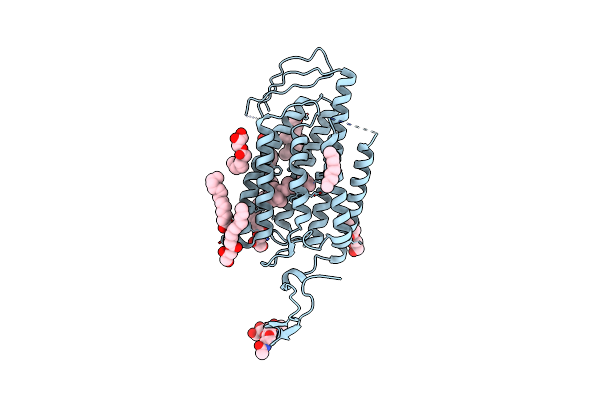 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 250 Microsecond Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
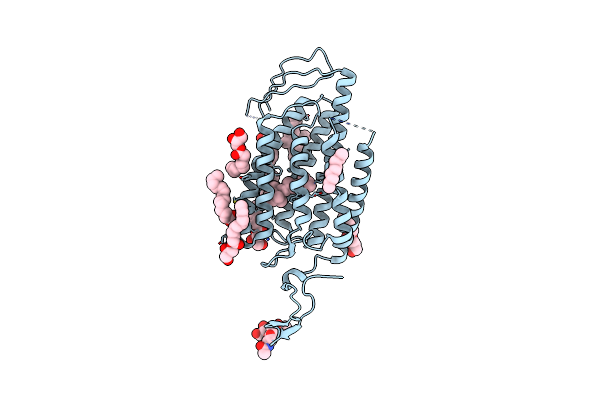 |
Time-Resolved Serial Femtosecond Crystallography Reveals Early Structural Changes In Channelrhodopsin: 1 Ms Structure
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-07 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
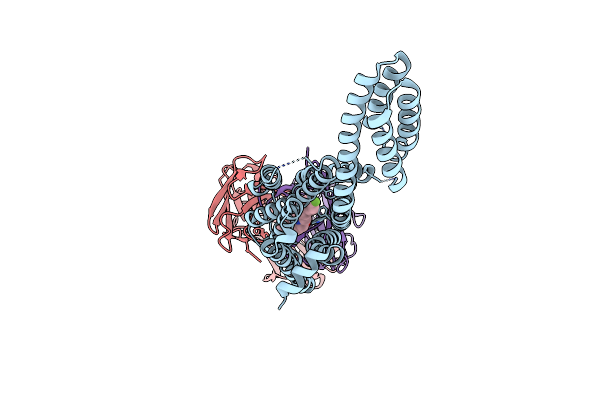 |
Organism: Homo sapiens, Escherichia coli, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-12-30 Classification: MEMBRANE PROTEIN Ligands: SIP |
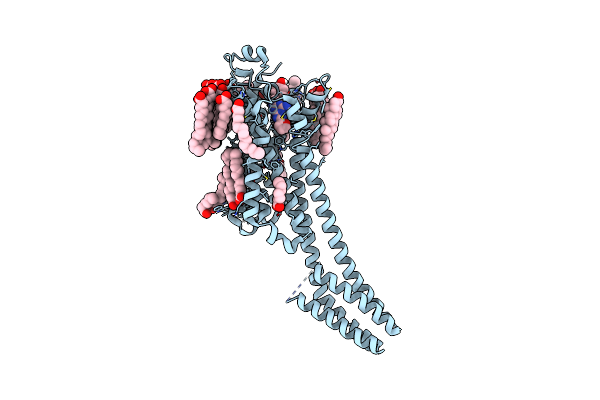 |
Structure Of Human A2A Adenosine Receptor In Complex With Zm241385 Obtained From Sfx Experiments Under Atmospheric Pressure
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-10-30 Classification: MEMBRANE PROTEIN |
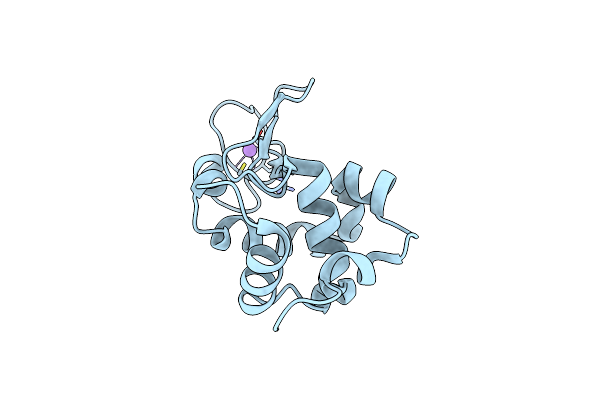 |
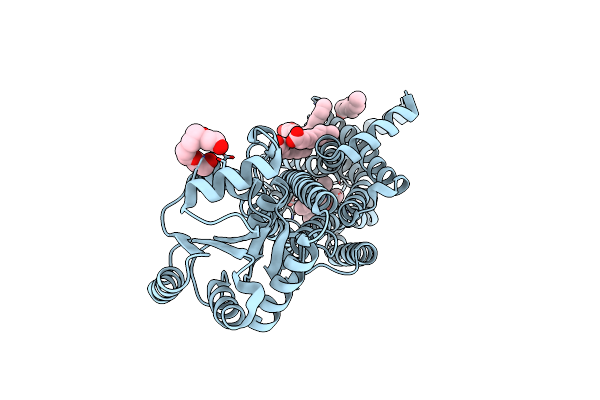
Structure Of Hen Egg-White Lysozyme Obtained From Sfx Experiments Under Atmospheric Pressure
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-10-30 Classification: HYDROLASE |
 |
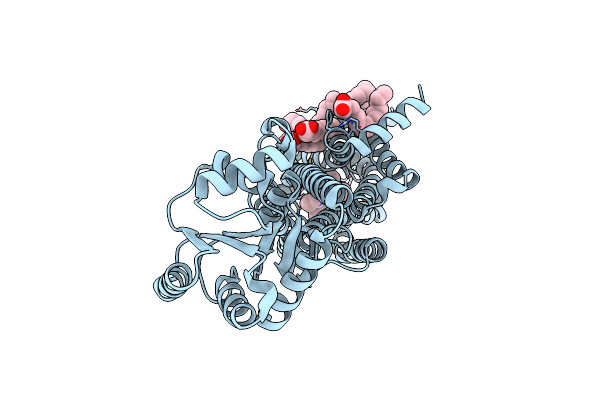
Crystal Structures Of Human Orexin 2 Receptor Bound To The Selective Antagonist Empa Determined By Serial Femtosecond Crystallography At Sacla
Organism: Homo sapiens, Pyrococcus abyssi (strain ge5 / orsay)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-12-13 Classification: SIGNALING PROTEIN Ligands: 7MA, OLA, 1PE |
 |
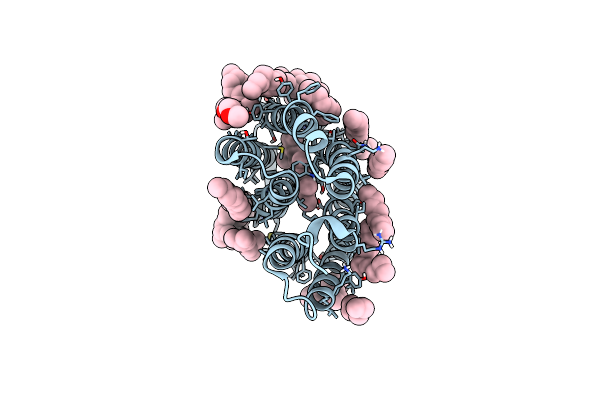
Crystal Structure Of Human Orexin 2 Receptor Bound To The Selective Antagonist Empa Determined By The Synchrotron Light Source At Spring-8.
Organism: Homo sapiens, Pyrococcus abyssi ge5
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2017-11-29 Classification: SIGNALING PROTEIN Ligands: 7MA, OLA, 1PE |
 |
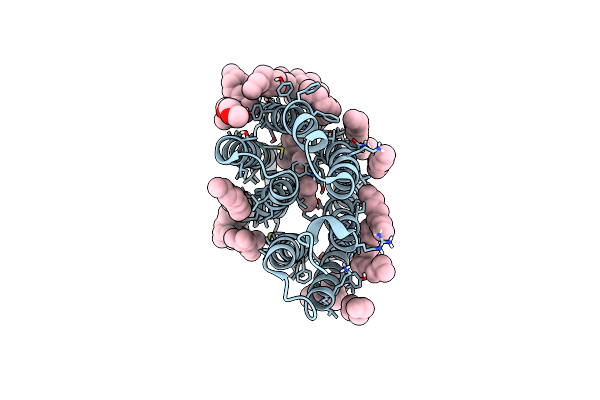
A Three Dimensional Movie Of Structural Changes In Bacteriorhodopsin: Resting State Structure
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-12-21 Classification: PROTON TRANSPORT Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
 |
A Three Dimensional Movie Of Structural Changes In Bacteriorhodopsin: Structure Obtained 16 Ns After Photoexcitation
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-12-21 Classification: PROTON TRANSPORT Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
 |
A Three Dimensional Movie Of Structural Changes In Bacteriorhodopsin: Structure Obtained 760 Ns After Photoexcitation
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-12-21 Classification: PROTON TRANSPORT Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |
 |
A Three Dimensional Movie Of Structural Changes In Bacteriorhodopsin: Structure Obtained 36.2 Us After Photoexcitation
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-12-21 Classification: PROTON TRANSPORT Ligands: RET, L2P, TRD, D10, HP6, OCT, MYS, UND, DD9, C14 |