Search Count: 37
 |
Crystal Structure Of Sars-Cov-2 Nucleocapsid Phosphoprotein N-Terminal Domain(N-Ntd) In Complex With Ump
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2025-04-23 Classification: VIRAL PROTEIN Ligands: U, PO4 |
 |
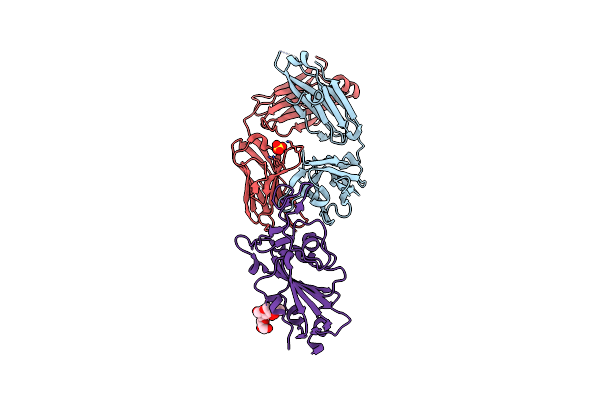
Crystal Structure Of Sars-Cov-2 Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg53 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-04-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: SO4, NAG |
 |
Crystal Structure Of Sars-Cov-2 Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg48 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2023-04-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: SO4 |
 |
Crystal Structure Of Sars-Cov-2 Omicron Variant Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg48 Fab
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2023-04-19 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: SO4 |
 |
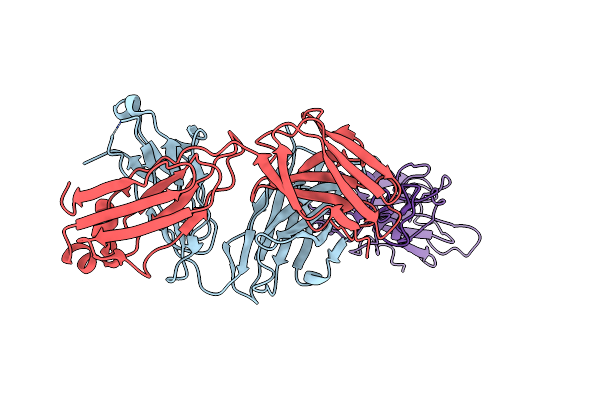
Crystal Structure Of Sars-Cov-2 Delta Variant Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg48 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2023-04-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
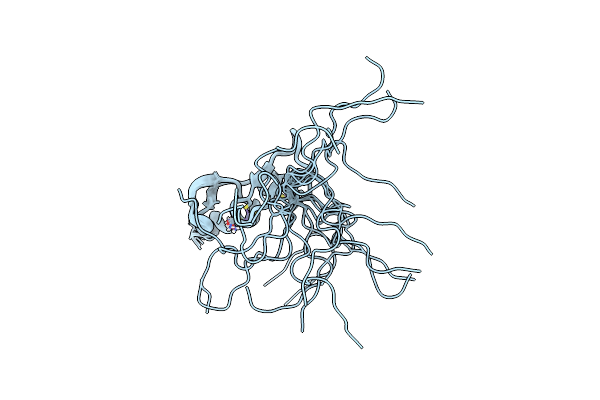
Crystal Structure Of Sars-Cov-2 Delta Variant Spike Receptor-Binding Domain (Rbd) In Complex With Ncv2Sg53 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2023-04-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2020-12-23 Classification: LIPID BINDING PROTEIN Ligands: ZN |
 |
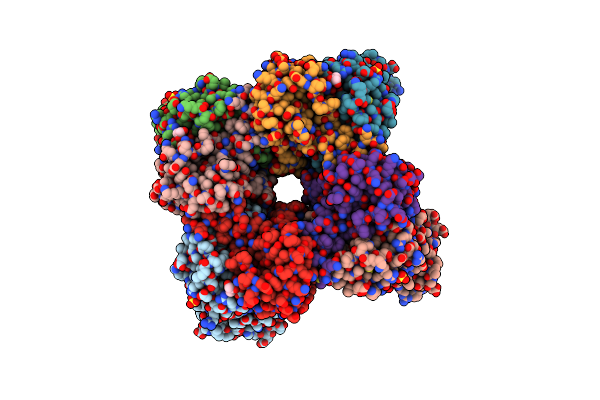
Organism: Oryza sativa, Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-09-16 Classification: PHOTOSYNTHESIS Ligands: SO4, GOL |
 |
Organism: Oryza sativa, Sorghum bicolor
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-09-16 Classification: PHOTOSYNTHESIS Ligands: SO4, GOL |
 |
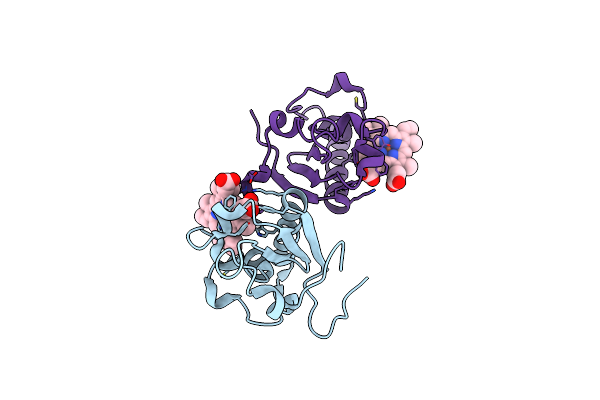
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With Ubiquinone-1
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, HEM, UQ1, EPH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-03-23 Classification: MEMBRANE PROTEIN Ligands: HEM |
 |
Crystal Structure Of Mitochondrial Rhodoquinol-Fumarate Reductase From Ascaris Suum With N-[3-(Pentafluorophenoxy)Phenyl]-2-(Trifluoromethyl)Benzamide
Organism: Ascaris suum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: MLI, FAD, FES, SF4, F3S, HEM, FD8 |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-06-15 Classification: LYASE Ligands: PLP, CL, ZN |
 |
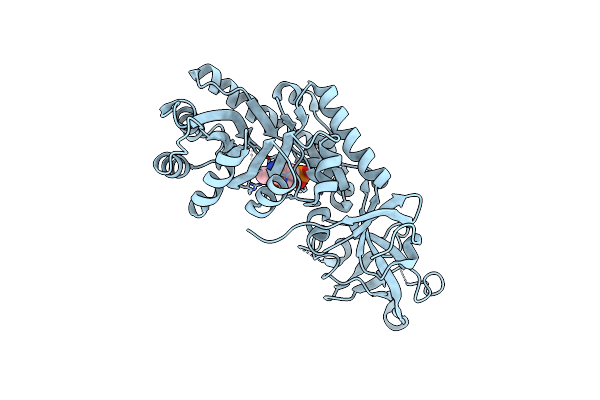
Crystal Structure Of D-Serine Dehydratase From Chicken Kidney (2,3-Dap Complex)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2011-06-15 Classification: LYASE Ligands: PLP, CL, ZN, 2RA |
 |
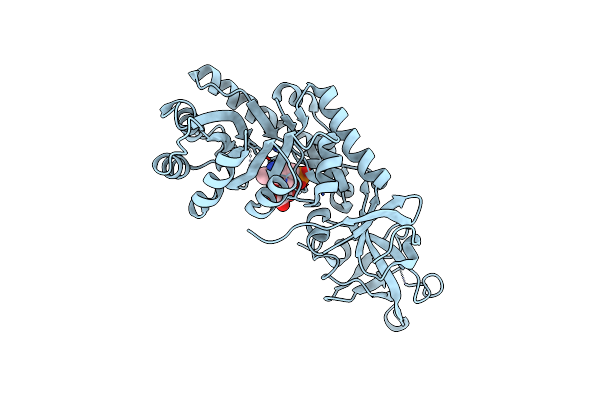
Crystal Structure Of D-Serine Dehydratase From Chicken Kidney (Edta Treated)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-06-15 Classification: LYASE Ligands: PLP |
 |
Crystal Structure Of D-Serine Dehydratase In Complex With D-Serine From Chicken Kidney (Edta-Treated)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2011-06-15 Classification: LYASE Ligands: PLP, DSN |
 |
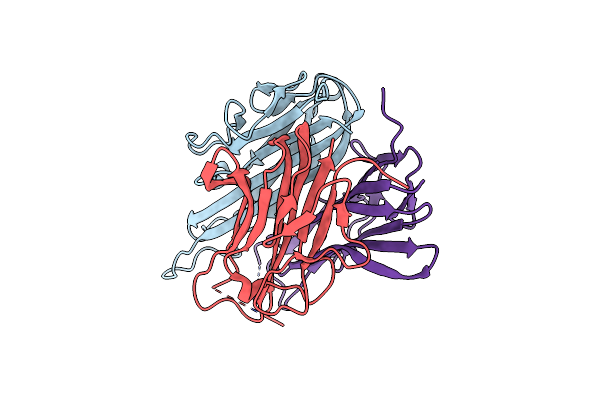
Crystal Structure Of The Met1-Form Of The Copper-Bound Tyrosinase In Complex With A Caddie Protein From Streptomyces Castaneoglobisporus Obtained By Soaking In Cupric Sulfate Solution For 36 Hours
Organism: Streptomyces castaneoglobisporus, Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2009-04-07 Classification: OXIDOREDUCTASE/METAL TRANSPORT Ligands: CU, NO3 |
 |
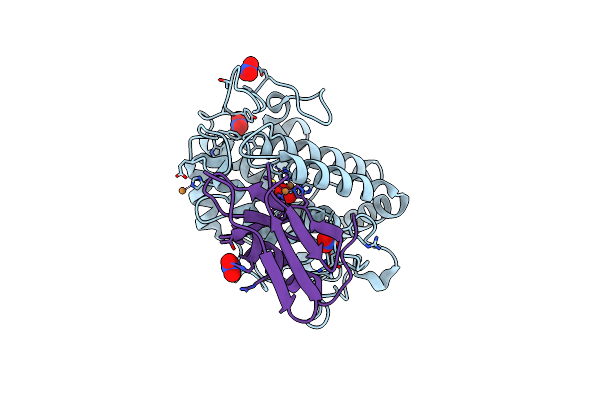
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-11-13 Classification: CYTOKINE |
 |
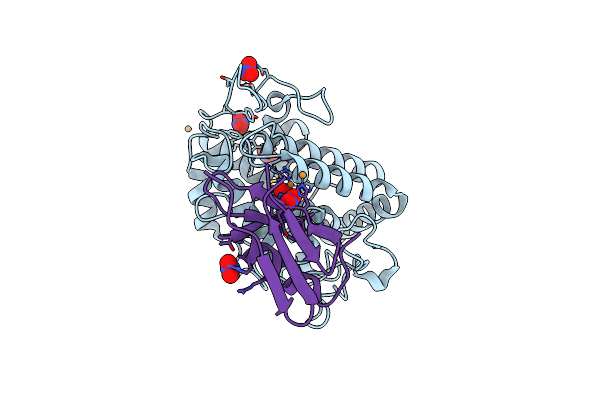
Crystal Structure Of The Oxy-Form Of The Copper-Bound Streptomyces Castaneoglobisporus Tyrosinase Complexed With A Caddie Protein Prepared By The Addition Of Hydrogenperoxide
Organism: Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-01-31 Classification: OXIDOREDUCTASE/METAL TRANSPORT Ligands: CU, NO3, PER |
 |
Crystal Structure Of The Oxy-Form Of The Copper-Bound Streptomyces Castaneoglobisporus Tyrosinase Complexed With A Caddie Protein Prepared By The Addition Of Dithiothreitol
Organism: Streptomyces castaneoglobisporus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2006-01-31 Classification: OXIDOREDUCTASE/METAL TRANSPORT Ligands: CU, NO3, PER |

