Search Count: 14
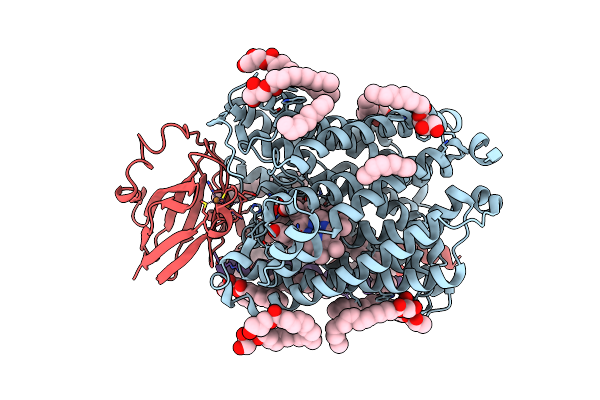 |
Serial Femtosecond Crystallography Structure Of Co Bound Ba3- Type Cytochrome C Oxidase Without Pump Laser Irradiation
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-15 Classification: ELECTRON TRANSPORT Ligands: CU, HEM, HAS, OLC, CMO, CUA |
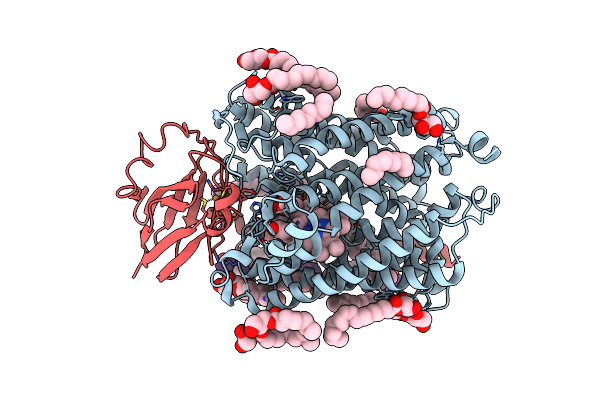 |
Serial Femtosecond Crystallography Structure Of Photo Dissociated Co From Ba3- Type Cytochrome C Oxidase Determined By Extrapolation Method
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-15 Classification: ELECTRON TRANSPORT Ligands: CU, HEM, HAS, OLC, CMO, CUA |
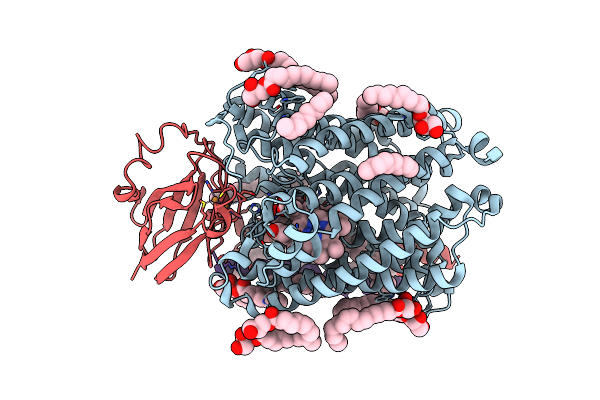 |
Serial Femtosecond Crystallography Structure Of Co Bound Ba3- Type Cytochrome C Oxidase At 2 Milliseconds After Irradiation By A 532 Nm Laser
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-08-16 Classification: ELECTRON TRANSPORT Ligands: CU, HEM, HAS, OLC, CMO, CUA |
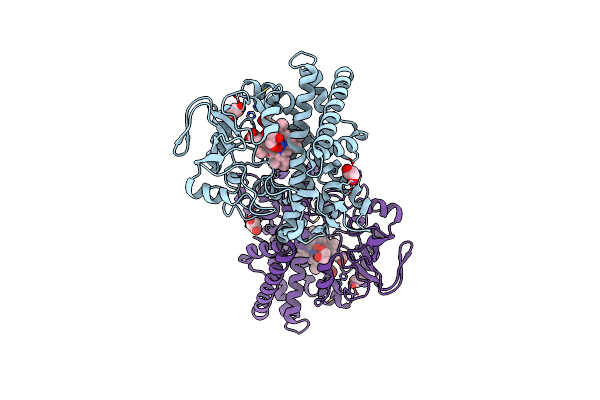 |
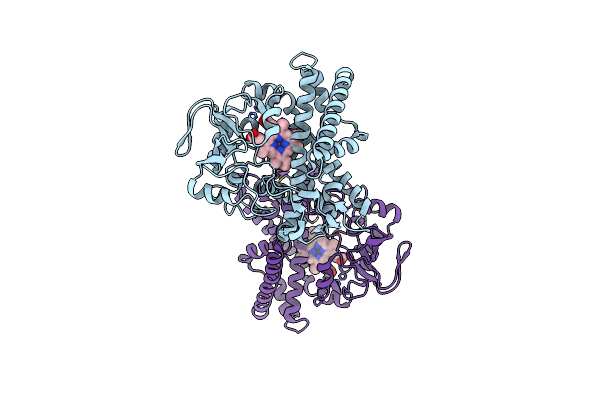
Structure Of Reaction Intermediate Of Cytochrome P450 No Reductase (P450Nor) Determined By Xfel
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-05-19 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
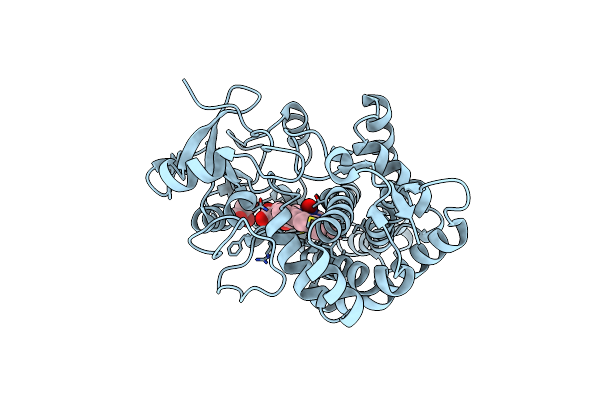
Sfx Structure Of Cytochrome P450Nor: A Complete Dark Data Without Pump Laser (Resting State)
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-08-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
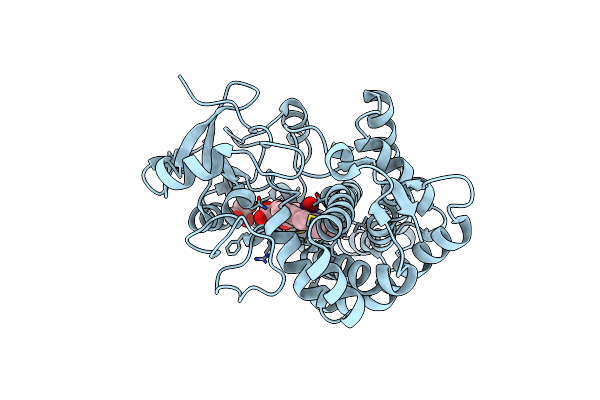
Structure Of Cytochrome P450Nor In No-Bound State: Damaged By Low-Dose (0.72 Mgy) X-Ray
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
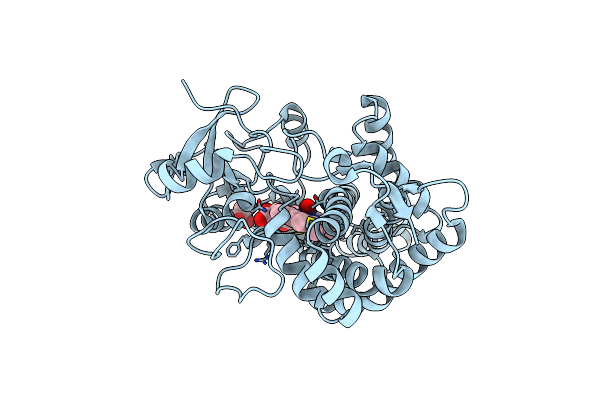
Structure Of Cytochrome P450Nor In No-Bound State: Damaged By High-Dose (5.7 Mgy) X-Ray
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
Sf-Rox Structure Of Cytochrome P450Nor (No-Bound State) Determined At Sacla
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
Time-Resolved Sfx Structure Of Cytochrome P450Nor: 20 Ms After Photo-Irradiation Of Caged No In The Presence Of Nadh (No-Bound State), Light Data
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM, NO, GOL |
 |
Time-Resolved Sfx Structure Of Cytochrome P450Nor: Dark-2 Data In The Presence Of Nadh (Resting State)
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM, GOL |
 |
Time-Resolved Sfx Structure Of Cytochrome P450Nor : 20 Ms After Photo-Irradiation Of Caged No In The Absence Of Nadh (No-Bound State), Light Data
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM, NO |
 |
Time-Resolved Sfx Structure Of Cytochrome P450Nor: Dark-2 Data In The Absence Of Nadh (Resting State)
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-12-06 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Complex Of Cytochrome Cd1 Nitrite Reductase And Nitric Oxide Reductase In Denitrification Of Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: HEC, CA, 10M, HEM, FE, O, DHE, CL |
 |
Cytochrome C-Dependent Nitric Oxide Reductase (Cnor) From Pseudomonas Aeruginosa In Complex With Xenon
Organism: Mus musculus, Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: HEM, FE, O, XE, 10M, CA, HEC |

