Search Count: 121
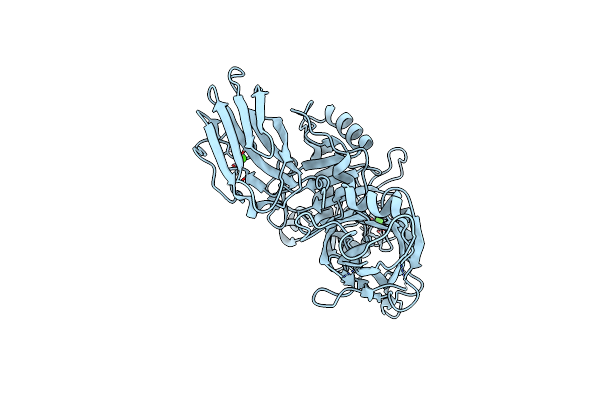 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
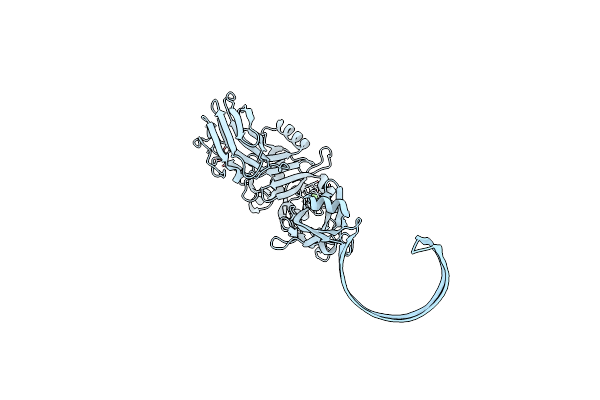 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TOXIN Ligands: CA |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: DNA Ligands: A1L1S |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: DNA Ligands: A1L1R |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: DNA Ligands: A1L1S |
 |
Organism: Clostridium perfringens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TOXIN Ligands: CA |
 |
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.36 Å, 1.86 Å Release Date: 2025-03-12 Classification: HYDROLASE |
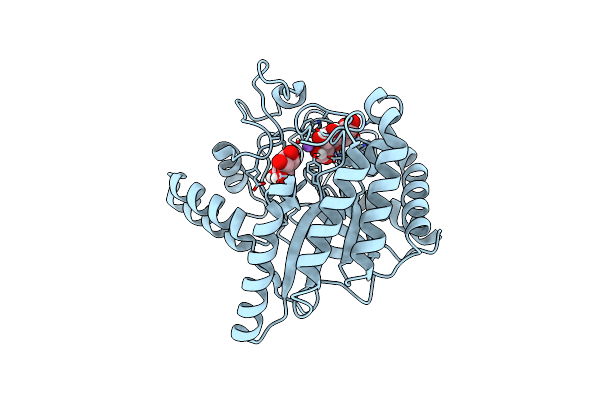 |
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature, Enzyme-Product Complex
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.40 Å, 1.8 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: BGC, NA |
 |
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature, Low-D2O-Solvent
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.40 Å, 2.15 Å Release Date: 2025-03-12 Classification: HYDROLASE |
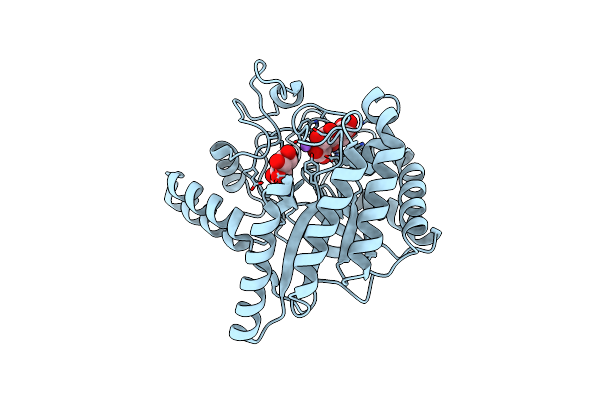 |
Neutron Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Room Temperature, Enzyme-Product Complex, H2O Solvent
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.80 Å, 2.59 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: BGC, NA |
 |
High-Resolution X-Ray Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Cryogenic Temperature
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION Resolution:0.80 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: MPD, ACT |
 |
High-Resolution X-Ray Structure Of Cellulase Cel6A From Phanerochaete Chrysosporium At Cryogenic Temperature, Enzyme-Product Complex
Organism: Phanerodontia chrysosporium
Method: X-RAY DIFFRACTION Resolution:0.85 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: BGC, NA, CL, PEG |
 |
Crystal Structure Of Trka D5 Domain In Complex With Two Different Macrocyclic Peptides
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-07-10 Classification: TRANSFERASE Ligands: EDO |

