Search Count: 8
 |
Structure Of Beta-1,2-Glucanase From Endozoicomonas Elysicola (Eesgl1, Ligand-Free)
Organism: Endozoicomonas elysicola dsm 22380
Method: X-RAY DIFFRACTION Release Date: 2025-01-22 Classification: HYDROLASE |
 |
Structure Of Beta-1,2-Glucanase From Photobacterium Gaetbulicola (Pgsgl3, Ligand-Free)
Organism: Photobacterium gaetbulicola gung47
Method: X-RAY DIFFRACTION Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CL |
 |
Structure Of Beta-1,2-Glucanase From Xanthomonas Campestris Pv. Campestris (Beta-1,2-Glucoheptasaccharide Complex)-E239Q Mutant
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Release Date: 2025-01-22 Classification: HYDROLASE |
 |
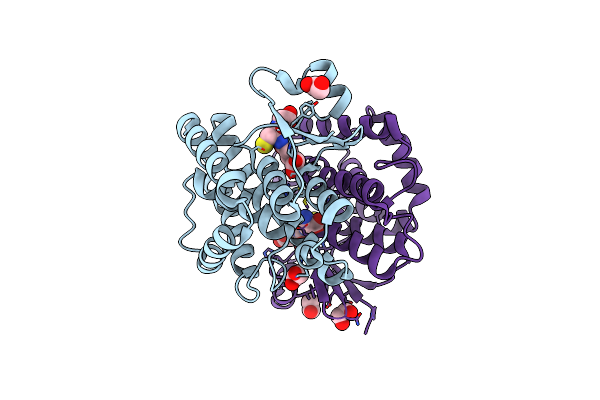
Crystal Structure Of A Diazinon-Metabolizing Glutathione S-Transferase In The Silkworm, Bombyx Mori
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-09-12 Classification: TRANSFERASE Ligands: EDO, ACT |
 |
Structural Characterization Of Catalytic Site Of A Nilaparvata Lugens Delta-Class Glutathione Transferase
Organism: Nilaparvata lugens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-01-14 Classification: TRANSFERASE Ligands: GSH, EDO |
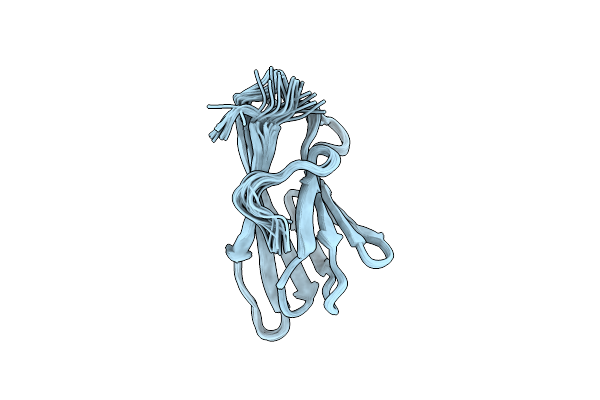 |
Solution Structure Of The Chitin-Binding Domain Of Chi18Ac From Streptomyces Coelicolor
Organism: Streptomyces coelicolor
Method: SOLUTION NMR Release Date: 2014-08-27 Classification: HYDROLASE |
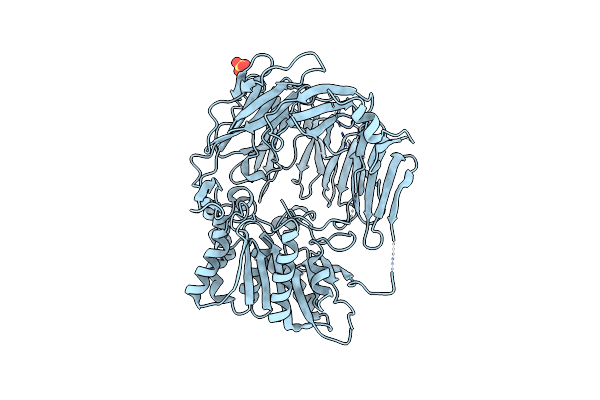 |
Crystal Structure Of Prolyl Tripeptidyl Aminopeptidase From Porphyromonas Gingivalis
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-09-19 Classification: HYDROLASE Ligands: SO4 |
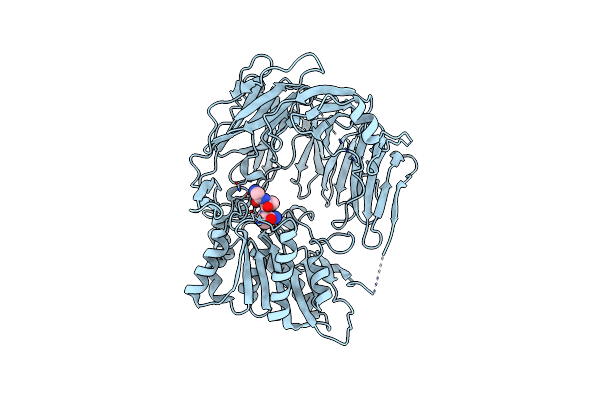 |
The Crystal Structure Of S603A Mutated Prolyl Tripeptidyl Aminopeptidase Complexed With Substrate
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2006-09-19 Classification: HYDROLASE Ligands: GA0 |

