Search Count: 17
 |
Crystal Structure Of An Abc Transporter Periplasmic Solute Binding Protein (Ipr025997) From Actinobacillus Succinogenes 130Z(Asuc_0081, Target Efi-511065) With Bound D-Allose
Organism: Actinobacillus succinogenes (strain atcc 55618 / 130z)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-10-07 Classification: TRANSPORT PROTEIN Ligands: ALL |
 |
Crystal Structure Of An Abc Transporter Solute Binding Protein From Agrobacterium Vitis(Avis_5339, Target Efi-511225) Bound With Alpha-D-Tagatopyranose
Organism: Agrobacterium vitis (strain s4 / atcc baa-846)
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2015-09-30 Classification: SUGAR BINDING PROTEIN Ligands: T6T |
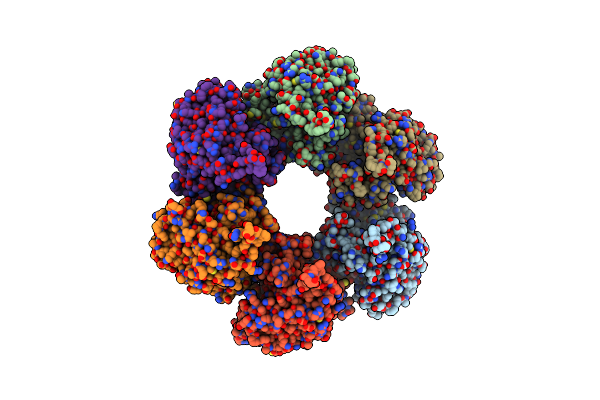 |
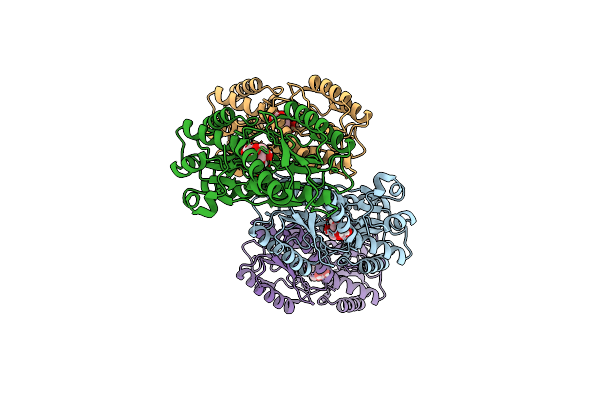
Crystal Structure Of Glutamine Synthetase From Chromohalobacter Salexigens Dsm 3043(Csal_0679, Target Efi-550015) With Bound Adp
Organism: Chromohalobacter salexigens (strain dsm 3043 / atcc baa-138 / ncimb 13768)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-09-16 Classification: LIGASE Ligands: ADP |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Pseudoalteromonas Atlantica T6C(Patl_2292, Target Efi-510180) With Bound Sodium And Pyruvate
Organism: Pseudoalteromonas atlantica
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2015-07-29 Classification: TRANSPORT PROTEIN Ligands: PYR, NA |
 |
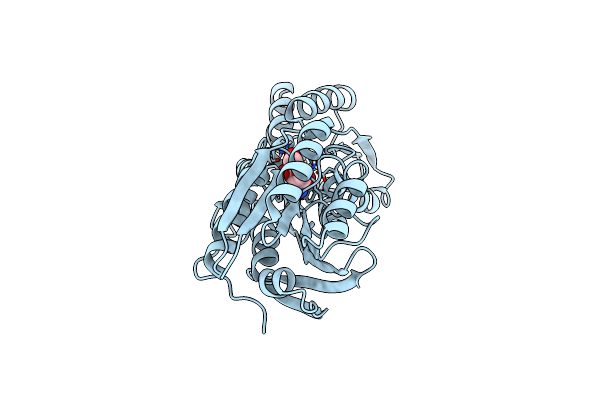
Crystal Structure Of An Abc Transporter Solute Binding Protein From Thermotoga Lettingae Tmo (Tlet_1705, Target Efi-510544) Bound With Alpha-D-Tagatose
Organism: Pseudothermotoga lettingae (strain atcc baa-301 / dsm 14385 / nbrc 107922 / tmo)
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2015-07-22 Classification: SUGAR BINDING PROTEIN Ligands: T6T, EDO, 1PE |
 |
Crystal Structure Of An Abc Transporter Solute Binding Protein (Ipr025997) From Agrobacterium Vitis S4 (Avi_5305, Target Efi-511224) With Bound Alpha-D-Galactosamine
Organism: Agrobacterium vitis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-06-10 Classification: SOLUTE-BINDING PROTEIN Ligands: X6X |
 |
Crystal Structure Of A Putative Periplasmic Solute Binding Protein (Ipr025997) From Ochrobactrum Anthropi Atcc49188 (Oant_2843, Target Efi-511085)
Organism: Ochrobactrum anthropi (strain atcc 49188 / dsm 6882 / nctc 12168)
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2015-06-10 Classification: SOLUTE-BINDING PROTEIN Ligands: HOH |
 |
Structure Of An Abc-Transporter Solute Binding Protein (Sbp_Ipr025997) From Actinobacillus Succinogenes (Asuc_0197, Target Efi-511067) With Bound Beta-D-Ribopyranose
Organism: Actinobacillus succinogenes (strain atcc 55618 / 130z)
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2015-05-20 Classification: SUGAR BINDING PROTEIN Ligands: RIP, EDO |
 |
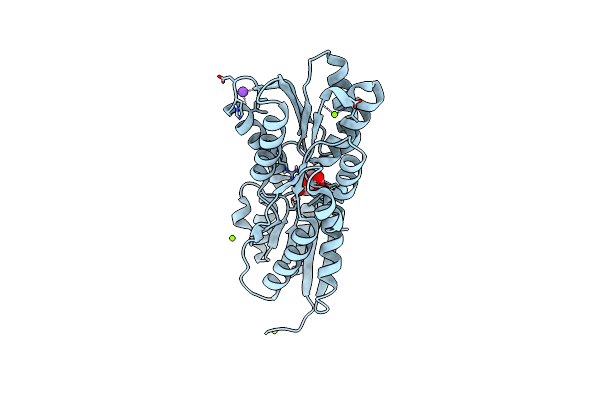
Crystal Structure Of A Periplasmic Solute Binding Protein (Ipr025997) From Streptobacillus Moniliformis Dsm-12112 (Smon_0317, Target Efi-511281) With Bound D-Galactose
Organism: Streptobacillus moniliformis
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2015-04-15 Classification: TRANSCRIPTION Ligands: SO4, GAL, ACT, CA, EDO, NA |
 |
Crystal Structure Of An Abc Transporter Solute Binding Protein (Ipr025997) From Bacillus Halodurans C-125 (Bh2323, Target Efi-511484) With Bound Myo-Inositol
Organism: Bacillus halodurans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-03-25 Classification: SOLUTE BINDING PROTEIN Ligands: MG, CL, INS, NA |
 |
Crystal Structure Of A Putative Dehydrogenase From Sulfitobacter Sp. (Cog1028) (Target Efi-513936) In Its Apo Form
Organism: Sulfitobacter sp. nas-14.1
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2015-03-25 Classification: OXIDOREDUCTASE Ligands: MG |
 |
Crystal Structure Of A Putative Oxidoreductase From Thermus Thermophilus Hb27 (Tt_P0034, Target Efi-513932) In Its Apo Form
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2015-03-25 Classification: OXIDOREDUCTASE Ligands: CL |
 |
Crystal Structure Of An Abc Transporter Solute Binding Protein (Ipr025997) From Agrobacterium Vitis S4 (Avi_5305, Target Efi-511224) With Bound Alpha-D-Glucosamine
Organism: Agrobacterium vitis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-11 Classification: SOLUTE-BINDING PROTEIN Ligands: PA1 |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Rhodobacter Sphaeroides (Rsph17029_2138, Target Efi-510205) With Bound Glucuronate
Organism: Rhodobacter sphaeroides (strain atcc 17029 / ath 2.4.9)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-12-24 Classification: SOLUTE-BINDING PROTEIN Ligands: BDP, PO4 |
 |
Crystal Structure Of A Tctc Solute Binding Protein From Polaromonas (Bpro_3516, Target Efi-510338), No Ligand
Organism: Polaromonas sp. (strain js666 / atcc baa-500)
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2014-12-24 Classification: UNKNOWN FUNCTION Ligands: CL |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Citrobacter Koseri (Cko_04899, Target Efi-510094) With Bound D-Glucuronate
Organism: Citrobacter koseri
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-12-10 Classification: SOLUTE BINDING PROTEIN Ligands: BDP, CL, MG |
 |
Crystal Structure Of A Putative D-Mannonate Oxidoreductase From Haemophilus Influenza (Avi_5165, Target Efi-513796) With Bound Nad
Organism: Haemophilus influenzae rdaw
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2014-11-26 Classification: OXIDOREDUCTASE Ligands: NAD, EDO |

