Search Count: 30
 |
Structure Of Duffy Antigen Receptor For Chemokines (Darc)/Ackr1 In Complex With The Chemokine, Ccl7 (Composite Map)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: IMMUNE SYSTEM |
 |
Organism: Xanthomonas campestris pv. campestris b100
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-05-01 Classification: HYDROLASE Ligands: GOL, ZN, NA |
 |
Crystal Structure Of M61 Peptidase (Bestatin-Bound) From Xanthomonas Campestris
Organism: Xanthomonas campestris pv. campestris b100
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-05-01 Classification: HYDROLASE Ligands: ZN, BES, GOL, TRS |
 |
Crystal Structure Of Escherichia Coli Adenine Phosphoribosyltransferase (Aprt) In Complex With Amp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-04-10 Classification: TRANSFERASE Ligands: AMP, CL, PO4, NA |
 |
Crystal Structure Of S9 Carboxypeptidase From Geobacillus Sterothermophilus
Organism: Geobacillus stearothermophilus atcc 12980
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-13 Classification: HYDROLASE Ligands: SO4, NA, GOL, ALA, FLC |
 |
Crystal Structure Of Escherichia Coli Adenine Phosphoribosyltransferase (Aprt) In Complex With Adenine
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-08-02 Classification: TRANSFERASE Ligands: ADE, ACT, CL, NA, EDO |
 |
Crystal Structure Of Escherichia Coli Adenine Phosphoribosyltransferase (Aprt) In Complex With Phosphate
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-04-12 Classification: TRANSFERASE Ligands: ACT, PO4, CL |
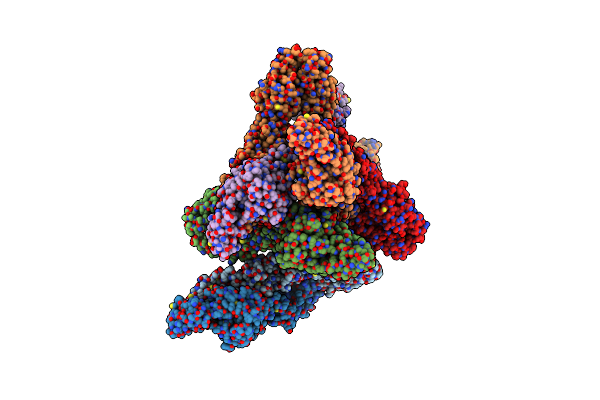 |
Organism: Eastern equine encephalitis virus
Method: ELECTRON MICROSCOPY Release Date: 2020-12-23 Classification: VIRUS |
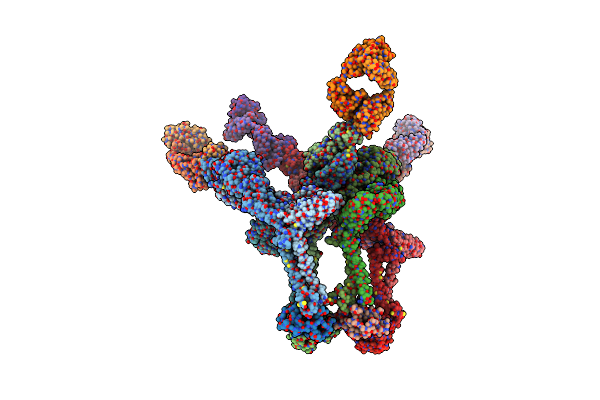 |
Cryoem Structure Of Eastern Equine Encephalitis (Eeev) Vlp With Fab Eeev-143.
Organism: Eastern equine encephalitis virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-23 Classification: Virus/Immune System |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2020-01-15 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
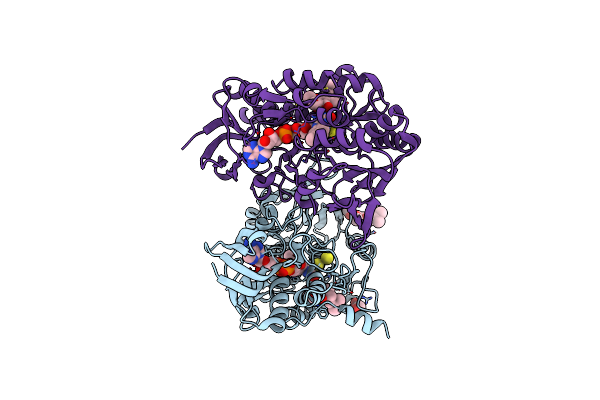
Crystal Structure Of Human Sulfide Quinone Oxidoreductase In Complex With Coenzyme Q (Sulfide Soaked)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2020-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, UQ1 |
 |
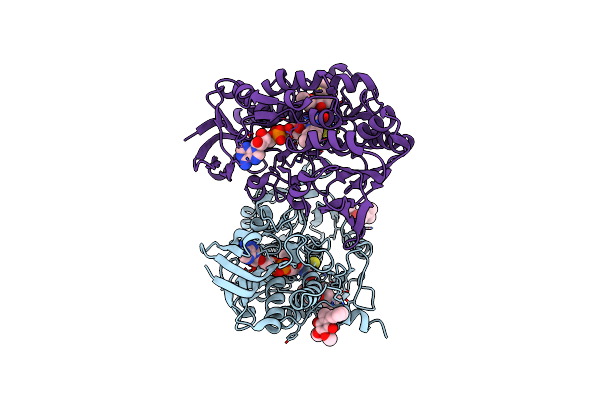
Crystal Structure Of Human Sulfide Quinone Oxidoreductase In Complex With Coenzyme Q
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2020-01-15 Classification: OXIDOREDUCTASE Ligands: GOL, FAD, UQ1, H2S |
 |
Crystal Structure Of Human Sulfide Quinone Oxidoreductase In Complex With Coenzyme Q (Sulfite Soaked)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2020-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, UQ1, H2S |
 |
Crystal Structure Of Peptidase E From Deinococcus Radiodurans In P6422 Space Group
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-11-20 Classification: HYDROLASE |
 |
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-26 Classification: HYDROLASE |
 |
Crystal Structure Of Human Cystathionine Gamma Lyase With S-3-Carboxpropyl-L-Cysteine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-06-12 Classification: LYASE Ligands: P1T |
 |
Crystal Structure Of S9 Peptidase (Inactive Form) From Deinococcus Radiodurans R1
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-14 Classification: HYDROLASE Ligands: ACT |
 |
Crystal Structure Of S9 Peptidase (Active Form) From Deinococcus Radiodurans R1
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-14 Classification: HYDROLASE |
 |
Crystal Structure Of S9 Peptidase Mutant (S514A) From Deinococcus Radiodurans R1
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-11-14 Classification: HYDROLASE Ligands: DMS, GOL |
 |
Crystal Structure Of S9 Peptidase (Inactive State)From Deinococcus Radiodurans R1 In P212121
Organism: Deinococcus radiodurans str. r1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-11-14 Classification: HYDROLASE Ligands: GOL |

