Search Count: 16
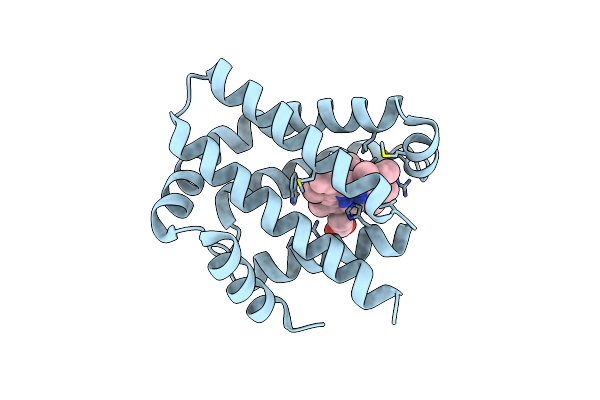 |
Crystal Structure Of The Globin Domain Of Thermosynechococcus Elongatus Bp-1
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-04-26 Classification: OXYGEN BINDING Ligands: IMD, HEM |
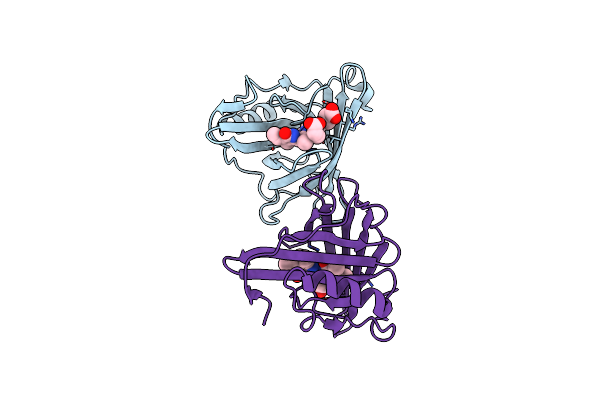 |
Organism: Sander
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-07-27 Classification: FLUORESCENT PROTEIN Ligands: BLA |
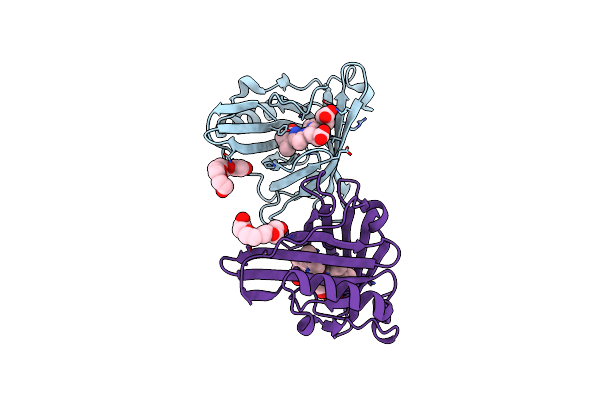 |
Organism: Sander vitreus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-05-25 Classification: FLUORESCENT PROTEIN Ligands: BLA, PG4 |
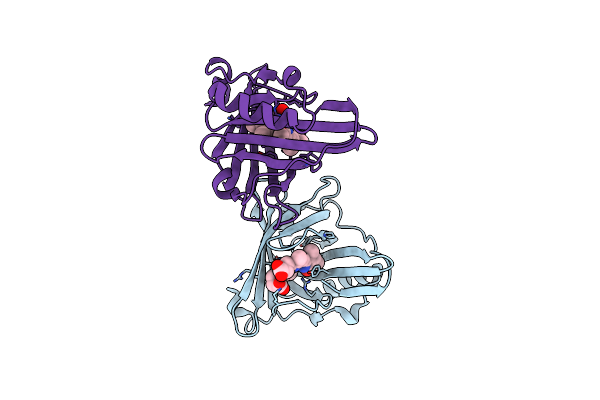 |
Organism: Sander vitreus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-05-25 Classification: FLUORESCENT PROTEIN Ligands: BLA |
 |
Organism: Sander vitreus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-04-13 Classification: FLUORESCENT PROTEIN Ligands: BLA |
 |
Structure Of A Monomeric Variant (L135E) Of Sandercyanin Fluorescent Protein Bound To Biliverdin Ix-Alpha
Organism: Sander vitreus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-04-13 Classification: FLUORESCENT PROTEIN Ligands: BLA |
 |
Cryo-Em Structure Of Nucleosome In Complex With A Single Chain Antibody Fragment
Organism: Drosophila melanogaster, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2019-05-22 Classification: NUCLEAR PROTEIN |
 |
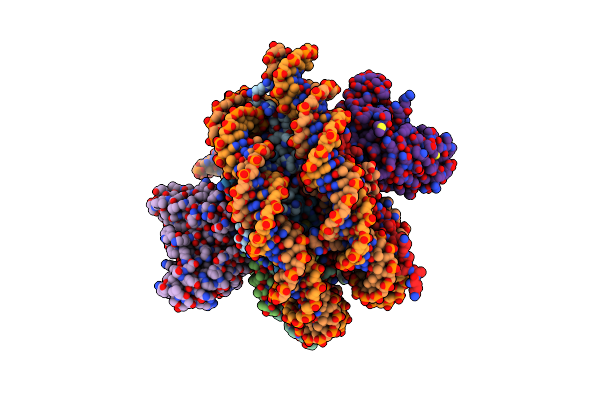
Cryo-Em Structure Of The Cenp-A Nucleosome (W601) In Complex With A Single Chain Antibody Fragment
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2019-05-22 Classification: NUCLEAR PROTEIN |
 |
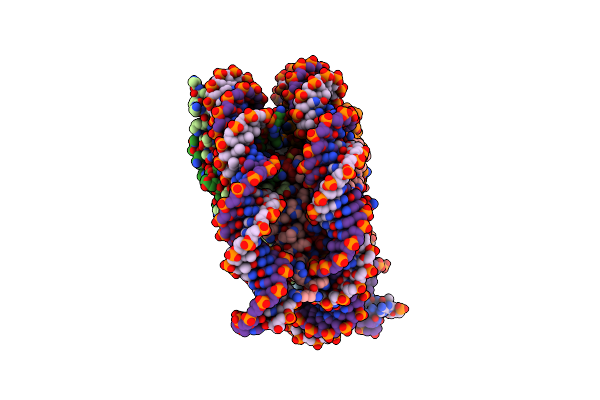
Cryo-Em Structure Of The Centromeric Nucleosome (Native Alpha Satellite Dna) In Complex With A Single Chain Antibody Fragment
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2019-05-22 Classification: NUCLEAR PROTEIN |
 |
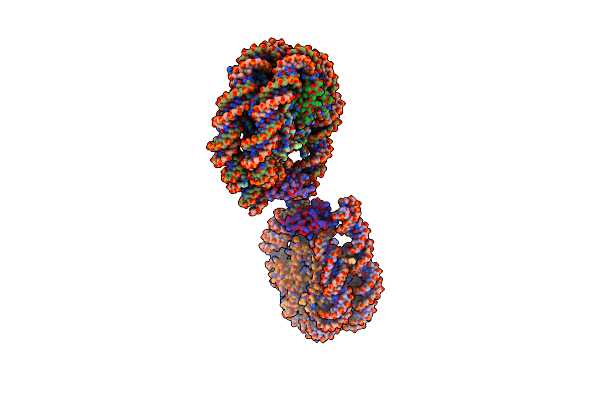
Cryo-Em Structure Of The Centromeric Nucleosome With Native Alpha Satellite Dna
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-05-22 Classification: NUCLEAR PROTEIN |
 |
Crystal Structure Of 167 Bp Nucleosome Bound To The Globular Domain Of Linker Histone H5
Organism: Drosophila melanogaster, Gallus gallus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:5.53 Å Release Date: 2018-10-31 Classification: CHROMATIN BINDING PROTEIN/DNA |
 |
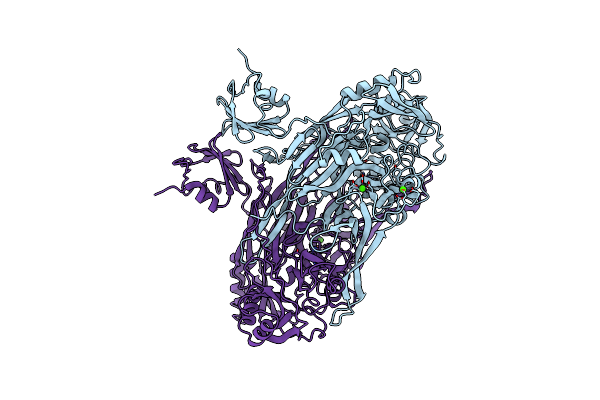
D-Aminoacyl-Trna Deacylase (Dtd) From Plasmodium Falciparum In Complex With Glycyl-3'-Aminoadenosine At 2.10 Angstrom Resolution
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: A3G |
 |
Crystal Structure Of A Quinoenzyme: Copper Amine Oxidase Of Escherichia Coli At 2 Angstroems Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1996-04-03 Classification: OXIDOREDUCTASE Ligands: CU, CA |
 |
Novel Thioether Bond Revealed By A 1.7 Angstroms Crystal Structure Of Galactose Oxidase
Organism: Hypomyces rosellus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1994-01-31 Classification: OXIDOREDUCTASE(OXYGEN(A)) Ligands: CU, NA, ACY |
 |
Novel Thioether Bond Revealed By A 1.7 Angstroms Crystal Structure Of Galactose Oxidase
Organism: Hypomyces rosellus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1994-01-31 Classification: OXIDOREDUCTASE(OXYGEN(A)) Ligands: CU, NA |
 |
Novel Thioether Bond Revealed By A 1.7 Angstroms Crystal Structure Of Galactose Oxidase
Organism: Hypomyces rosellus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1994-01-31 Classification: OXIDOREDUCTASE(OXYGEN(A)) |

