Search Count: 86
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: VIRAL PROTEIN |
 |
Organism: Oryza sativa subsp. japonica
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: TRANSPORT PROTEIN |
 |
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Organism: Weissella ceti
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: PG4 |
 |
Crystal Structure Of Trehalose-6-Phosphate Phosphorylase From Weissella Ceti In Complex With Beta-Glc1P
Organism: Weissella ceti
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: XGP, MG |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: LYASE |
 |
Structure Of Oleate Hydratase Mutant - V135A/L212V From Staphylococcus Aureus In The Complex With Fad
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: LYASE Ligands: FAD |
 |
Structure Of Oleate Hydratase Mutant L151V From Staphylococcus Aureus In The Complex With Linoleic Acid
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: LYASE Ligands: EIC, PEG, GOL |
 |
Crystal Structure Of Open State Ferulic Acid Decarboxylase From Saccharomyces Cerevisiae, F397V/I398L/T438P/P441V Mutant
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2025-01-01 Classification: LYASE Ligands: GOL, PEG |
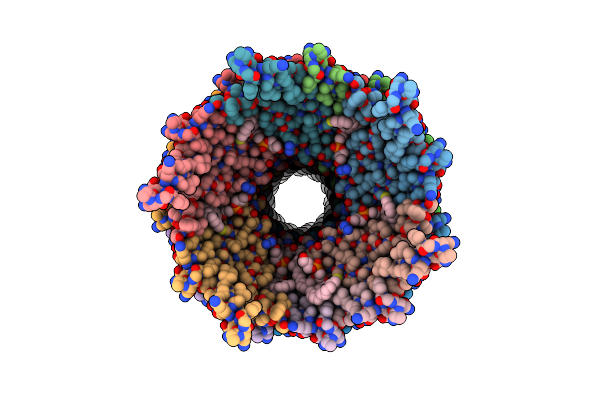 |
Cryo-Em Structure Of The R388 Plasmid Conjugative Pilus Reveals A Helical Polymer Characterised By An Unusual Pilin/Phospholipid Binary Complex
Organism: Escherichia coli hb101
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: PROTEIN FIBRIL Ligands: LHG |
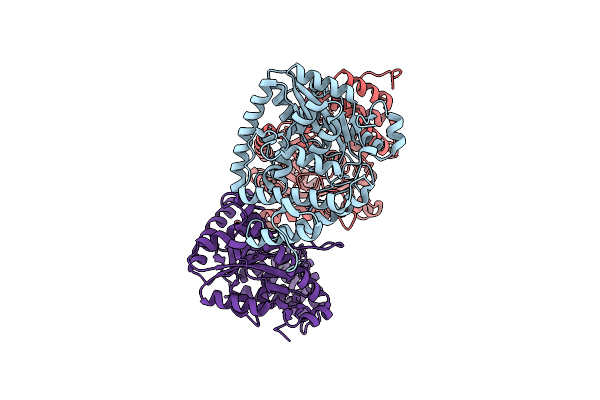 |
Plastidial Glycerol-3-Phosphate Acyltransferases (Gpat) From The Green Alga Myrmecia Incisa
Organism: Lobosphaera incisa
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2024-02-07 Classification: TRANSFERASE |
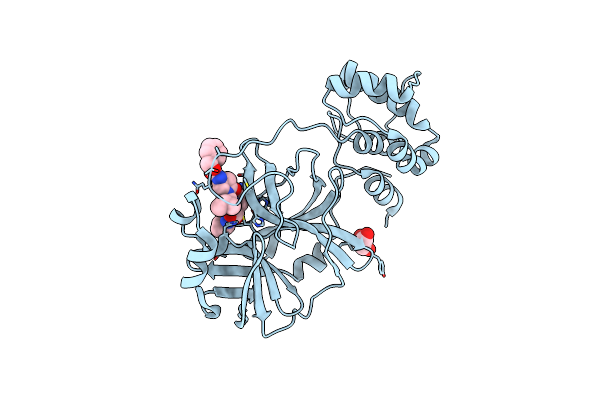 |
Crystal Structure Of The Sars-Cov-2 (Covid-19) Main Protease In Complex With Inhibitor 11
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-04-05 Classification: VIRAL PROTEIN/INHIBITOR Ligands: GOL, U6Y |
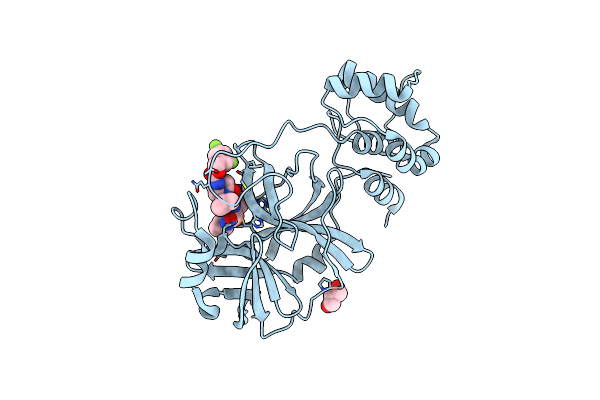 |
Crystal Structure Of The Sars-Cov-2 (Covid-19) Main Protease In Complex With Inhibitor 17
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-04-05 Classification: VIRAL PROTEIN/INHIBITOR Ligands: U76, GOL |
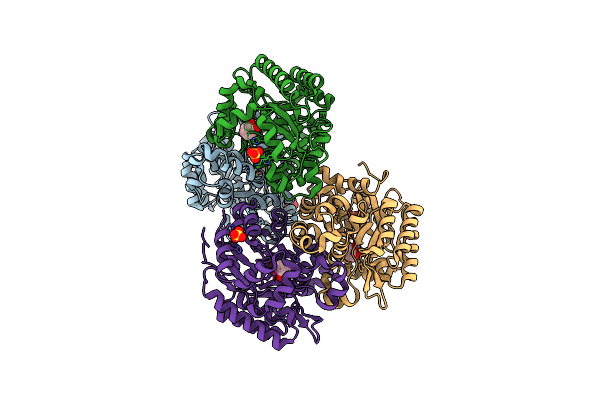 |
Crystal Structure Of Dihydroxybenzoate Decarboxylase Mutant F296Y From Aspergillus Oryzae In Complex With Catechol
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: MG, CAQ, SO4 |
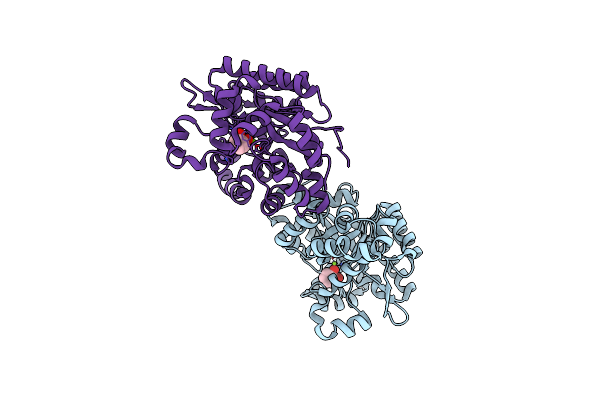 |
Crystal Structure Of 2,3-Dihydroxybenzoate Decarboxylase Complexed With Catechol
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: MG, CAQ |
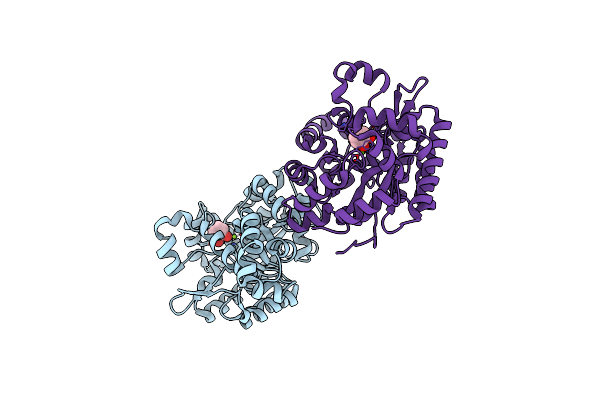 |
Crystal Structure Of 2,3-Dihydroxybenzoate Decarboxylase Mutant W23Y From Aspergillus Oryzae In Complex With Catechol
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: MG, CAQ |
 |
Crystal Structure Of Nicotinic Acid Mononucleotide Adenylyltransferase Mutant P22K/Y84V/Y118D/C132Q/W176F From Escherichia Coli
Organism: Escherichia coli nccp15648
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2020-07-15 Classification: TRANSFERASE |
 |
Crystal Structure Of Nicotinic Acid Mononucleotide Adenylyltransferase Mutant P22K/Y84V/Y118D/C132L/W176Y From Escherichia Coli
Organism: Escherichia coli nccp15648
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2020-07-15 Classification: LYASE |
 |
Crystal Structure Of Alcohol Dehydrogenase From Geobacillus Stearothermophilus
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2019-06-05 Classification: OXIDOREDUCTASE Ligands: ZN |
 |
Organism: Ralstonia sp. 4506
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-03-13 Classification: OXIDOREDUCTASE |

