Search Count: 632
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-28 Classification: DE NOVO PROTEIN Ligands: BLA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2026-01-28 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2026-01-28 Classification: DE NOVO PROTEIN |
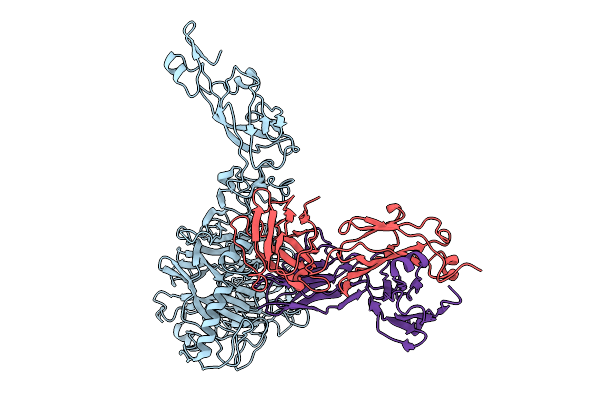 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: PROTEIN BINDING/IMMUNE SYSTEM |
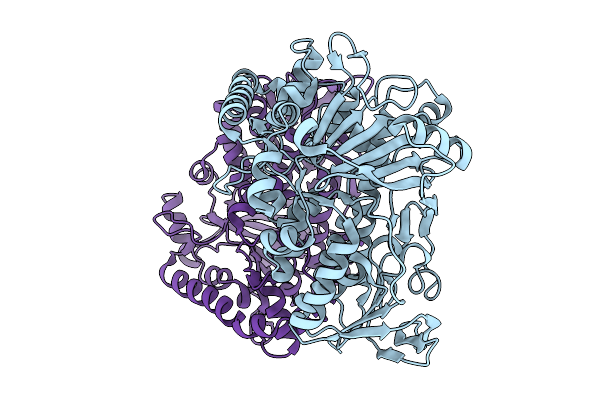 |
Organism: Nocardiopsis flavescens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2026-01-21 Classification: BIOSYNTHETIC PROTEIN |
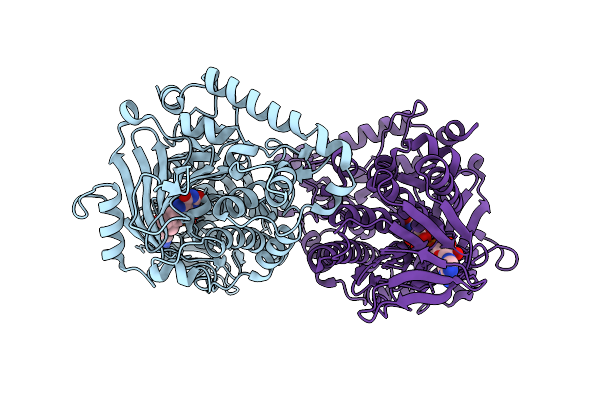 |
Organism: Nocardiopsis flavescens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2026-01-21 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD |
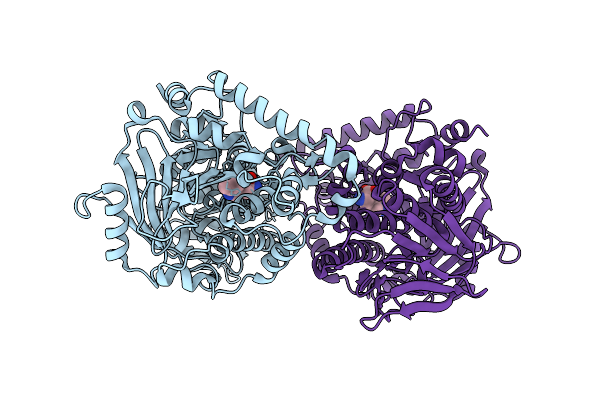 |
Organism: Nocardiopsis flavescens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2026-01-21 Classification: BIOSYNTHETIC PROTEIN Ligands: TRP |
 |
Organism: Nocardiopsis flavescens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-21 Classification: BIOSYNTHETIC PROTEIN Ligands: A1L7E |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:2.84 Å Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: CLR, A1EK5 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.62 Å Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: 91Q, CLR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.76 Å Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: A1EK5, CLR |
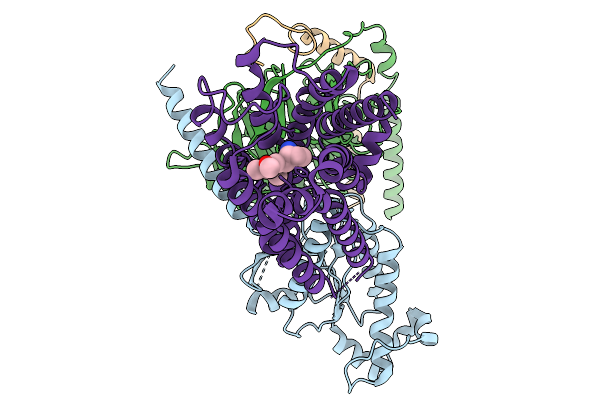 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.27 Å Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: A1EK8 |
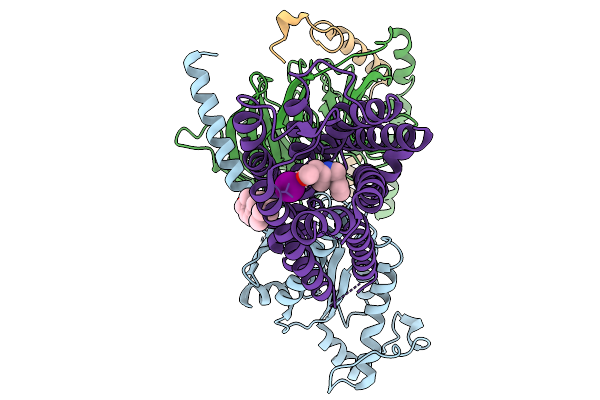 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.98 Å Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: CLR, A1ELA |
 |
The 1:1 Cryo-Em Structure Of Bap1/Asxl1-K351Ub In Complex With H2Ak119Ub Nucleosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: NUCLEAR PROTEIN/DNA |
 |
Organism: Glycyrrhiza inflata
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2026-01-07 Classification: BIOSYNTHETIC PROTEIN |
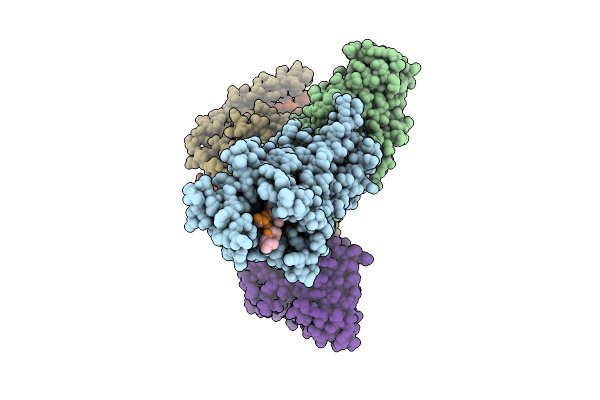 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.02 Å Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN |
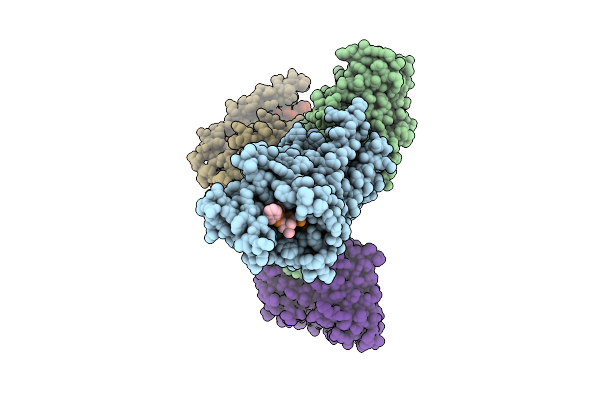 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.19 Å Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:3.13 Å Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: 8K3 |
 |
Organism: Burkholderia cenocepacia, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |

