Search Count: 954
 |
Organism: Homo sapiens, Rattus norvegicus, Bos taurus, Mus musculus, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: 8WL |
 |
Organism: Homo sapiens, Rattus norvegicus, Bos taurus, Lama glama, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: DR7 |
 |
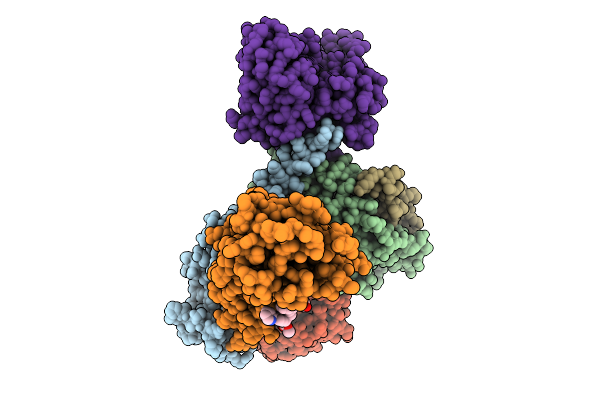
Structure Of Beta1-Ar-Gs Complex Bound To Epinephrine And An Allosteric Modulator
Organism: Bos taurus, Rattus norvegicus, Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: DR7, ALE |
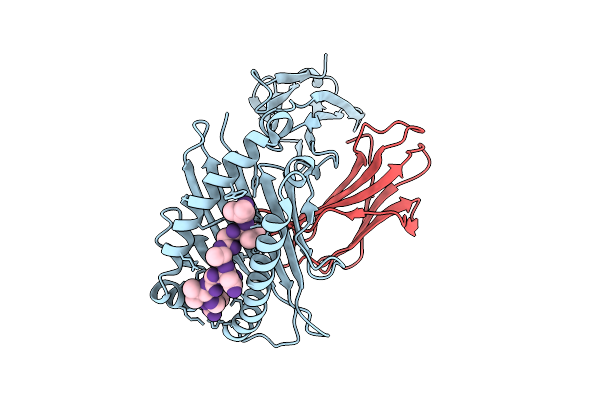 |
Crystal Structure Of Hla-A*11:01 In Complex With Kras G12D 9-Mer Peptide (Vvgadgvgk)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
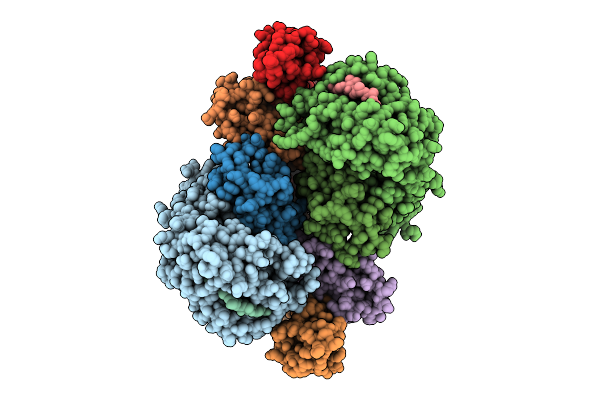 |
Crystal Structure Of Hla-A*11:01 In Complex With Kras G12A 10-Mer Peptide (Vvvgaagvgk)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
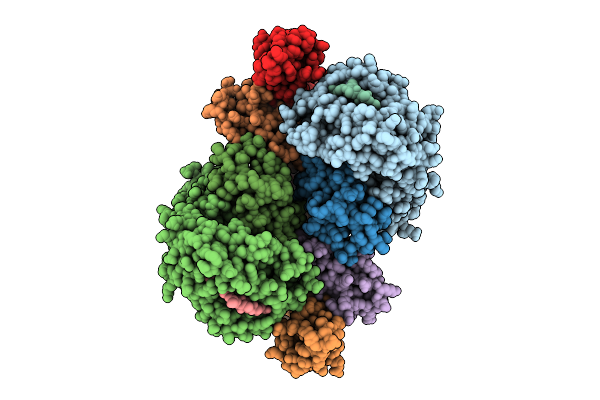 |
Crystal Structure Of Hla-A*11:01 In Complex With Kras G12S 10-Mer Peptide (Vvvgasgvgk)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM |
 |
Mycobacterium Tuberculosis Relbe1 Toxin-Antitoxin System; Rv1247C (Relb1 Antitoxin), Rv1246C (Rele1 Toxin)
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TOXIN Ligands: CL |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: ARG, HEM |
 |
Organism: Streptomyces monomycini
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, ARG, PGL |
 |
Cryo-Em Structure Of [Pen5]-Urotensin (4-11)-Bounded Human Urotensin Receptor (Uts2R)-Gq Complex
Organism: Homo sapiens, Lama glama, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: SIGNALING PROTEIN Ligands: CHO |
 |
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: GUN, MG |
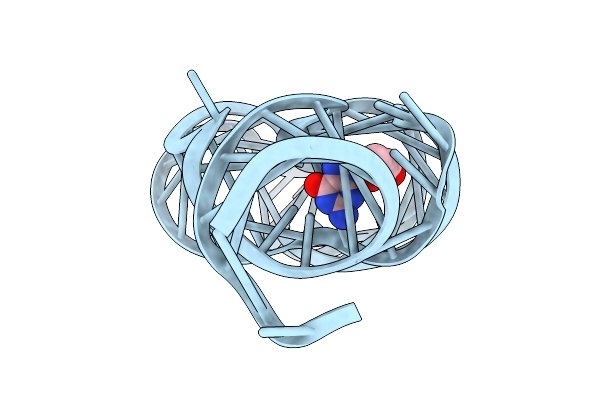 |
Crystal Structure Of Guanine-Ii Riboswitch In Complex With 2'-Deoxyguanosine
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: MG, GNG |
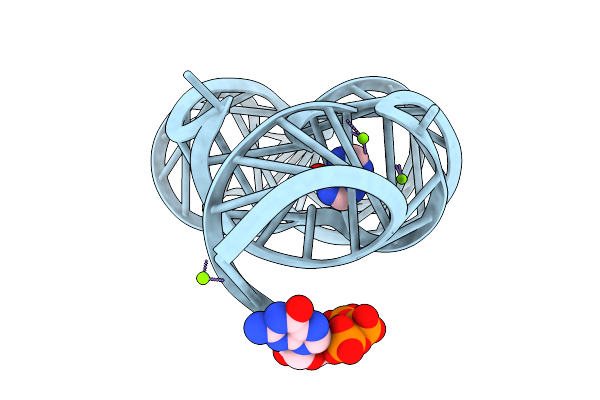 |
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: HPA, GTP, MG |
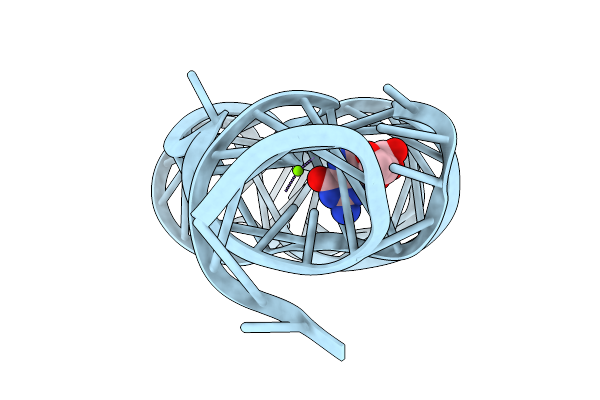 |
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: GMP, MG |
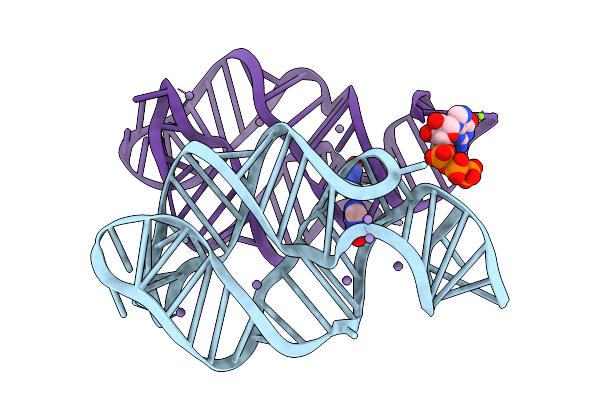 |
Crystal Structure Of Guanine-Ii Riboswitch In Complex With Guanine Soaked With Mn2+
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: GUN, MN, GTP, MG |
 |
Crystal Structure Of Guanine-Ii Riboswitch In Complex With 7,8-Dihydroneopterin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: NPR, MG |

