Search Count: 748
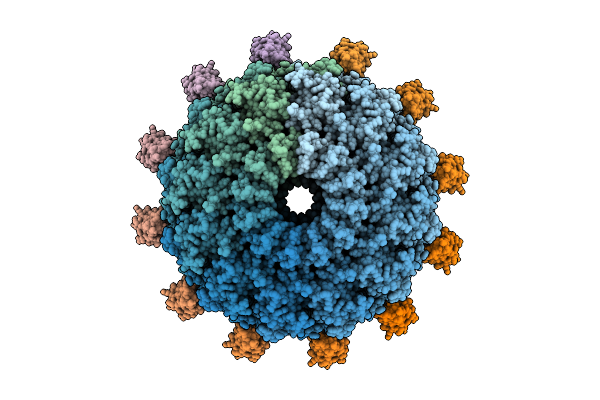 |
Organism: Oshimavirus p7426
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRUS |
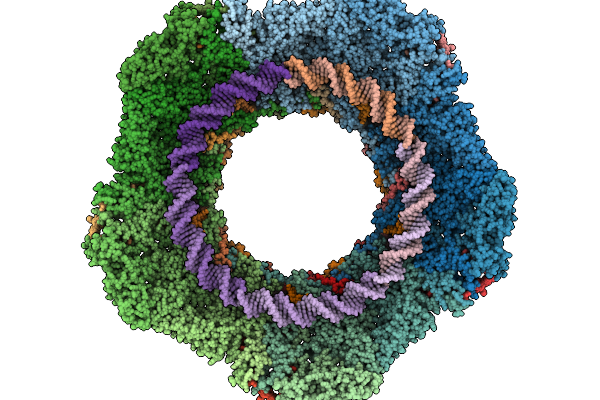 |
Organism: Oshimavirus p7426
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRUS |
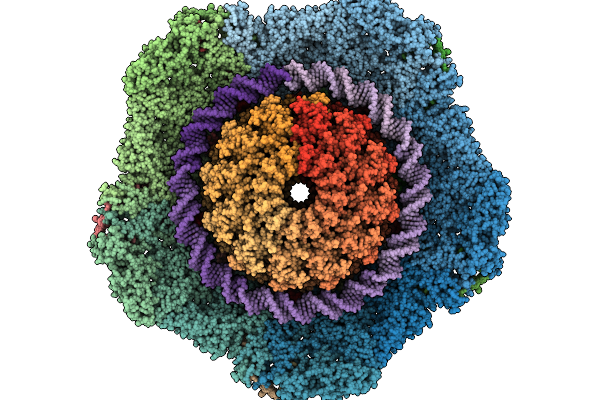 |
C1 Reconstruction Of The Thermophilic Bacteriophage P74-26 Portal And Portal Vertex
Organism: Oshimavirus p7426
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRUS |
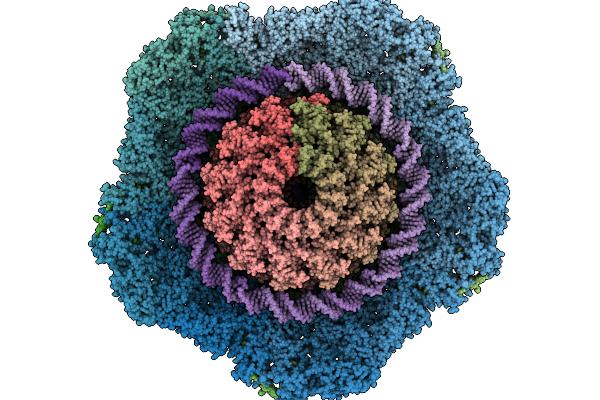 |
Composite Reconstruction Of The Thermophilic Bacteriophage P74-26 Neck And Portal Vertex
Organism: Oshimavirus p7426
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRUS/DNA |
 |
Organism: Oshimavirus p7426
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRUS/DNA |
 |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: CDL, PEE, NDP, ZMP, ADP, 3PE, PC1, PLX, XEW, SF4, FES, MG, ZN, FMN |
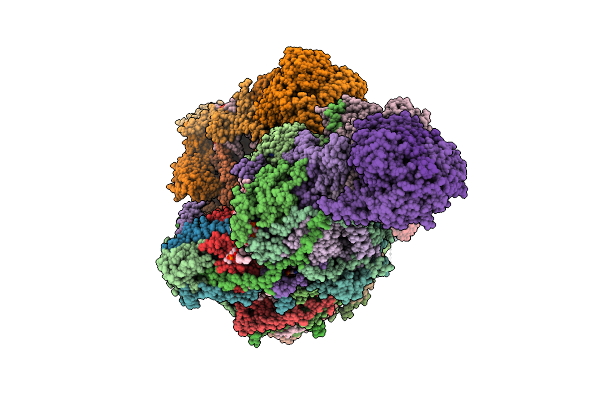
 |
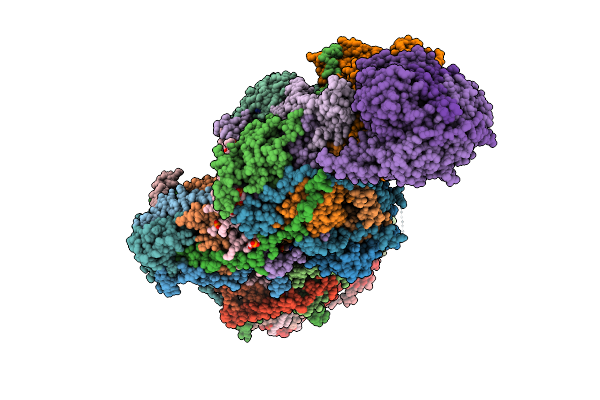
 |
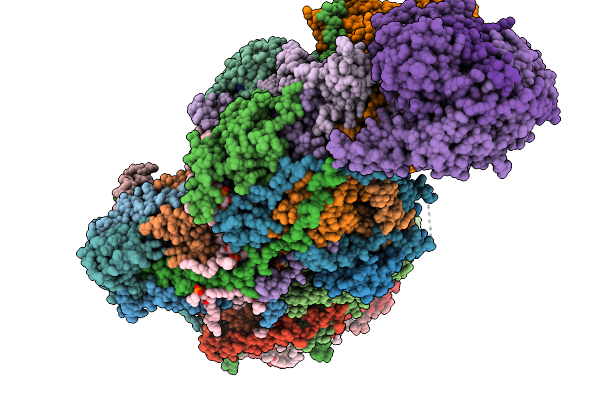 |
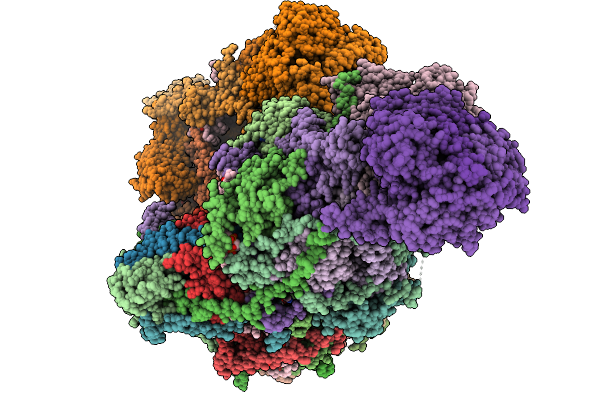
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10, Alternative Orientation (Sc-Metc1-Ii)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, PC1, NDP, PEE, ZMP, ADP, PLX, 3PE, PX2, U10, SF4, FES, MG, MF8, ZN, FMN |
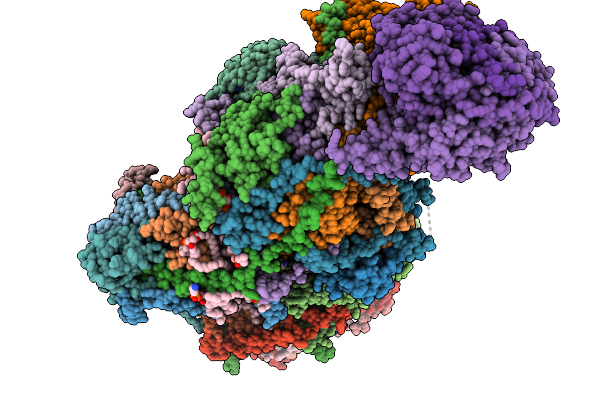 |
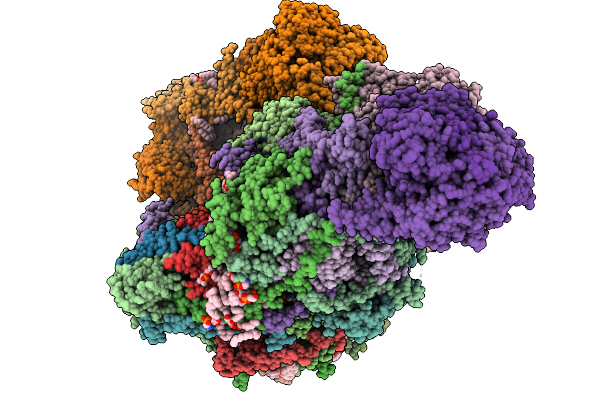
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10 (Sc-Metc1-I)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, PC1, NDP, PEE, ZMP, ADP, PLX, 3PE, U10, SF4, FES, MG, MF8, ZN, FMN |
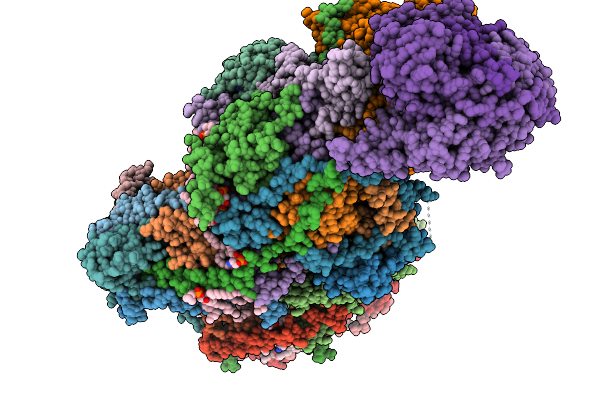 |
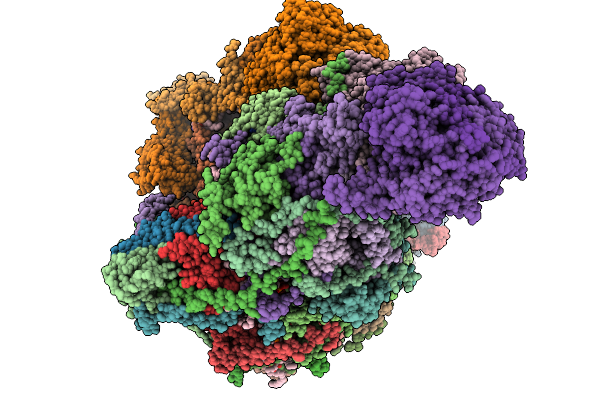
Complex I From Respirasome Closed State 1 Bound By Metformin (Sc-Metc1-Iii)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, PC1, NDP, PEE, ZMP, ADP, PLX, 3PE, SF4, FES, MG, MF8, ZN, FMN |
 |
 |
 |
 |
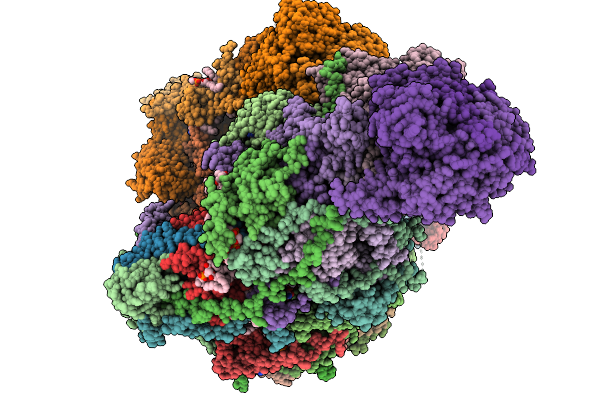
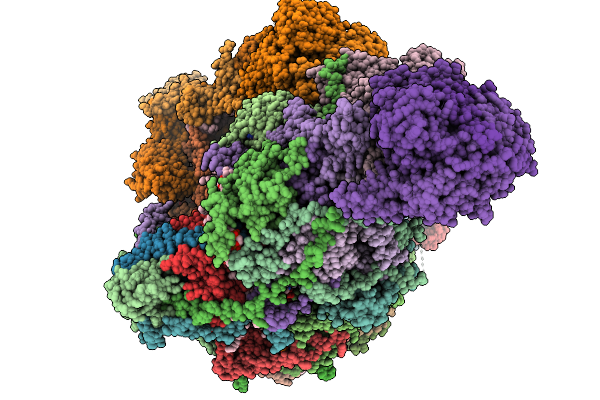
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, NDP, PEE, ZMP, ADP, PLX, 3PE, XEW, 6F6, SF4, FES, MG, ZN, FMN |
 |
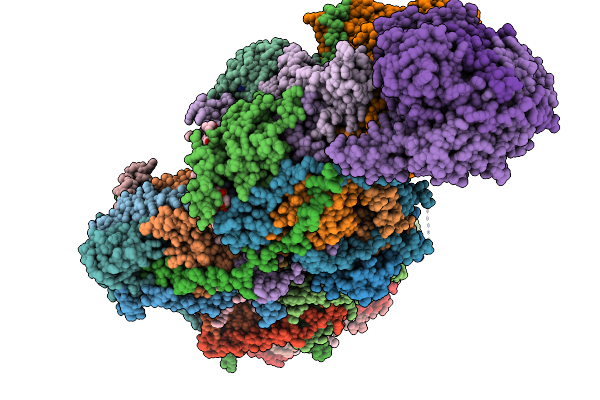
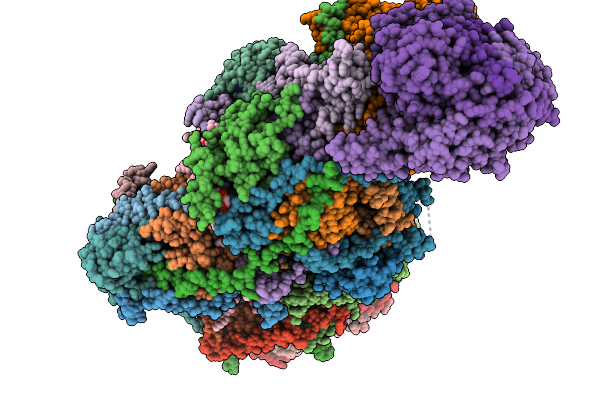
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10 (Sc-Metc1-Iv)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, NDP, PEE, PC1, ZMP, ADP, PLX, 3PE, U10, SF4, FES, MG, MF8, ZN, FMN |
 |
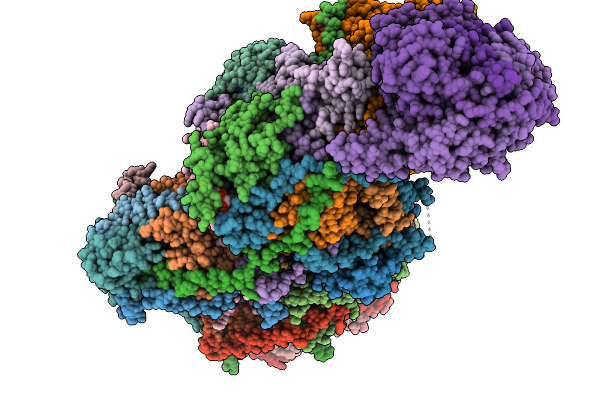
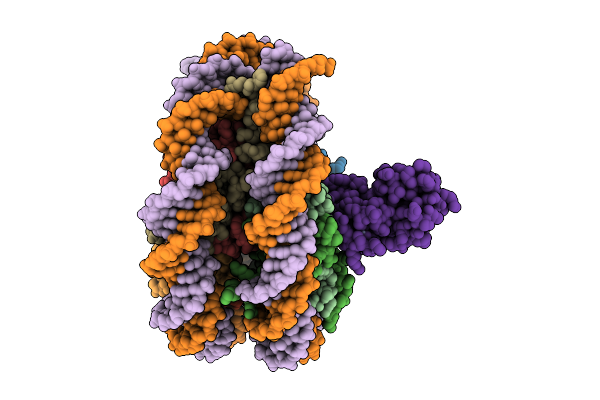
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10 (Sc-Metc1-V)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, NDP, PEE, ZMP, ADP, PLX, 3PE, PC1, SF4, FES, MG, ZN, U10, MF8, FMN |
 |
Organism: Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
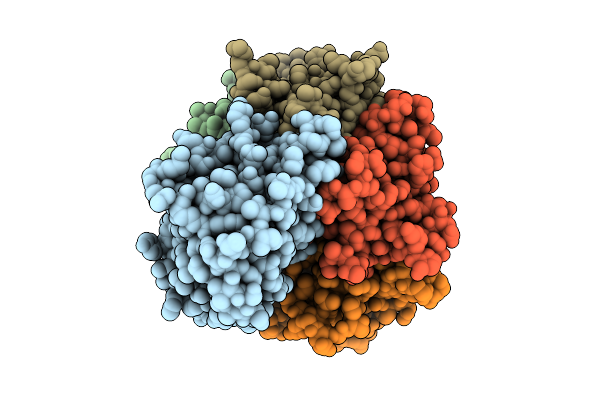 |
Organism: Salmonella enterica subsp. diarizonae
Method: ELECTRON MICROSCOPY Resolution:2.32 Å Release Date: 2025-10-29 Classification: TOXIN |

