Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
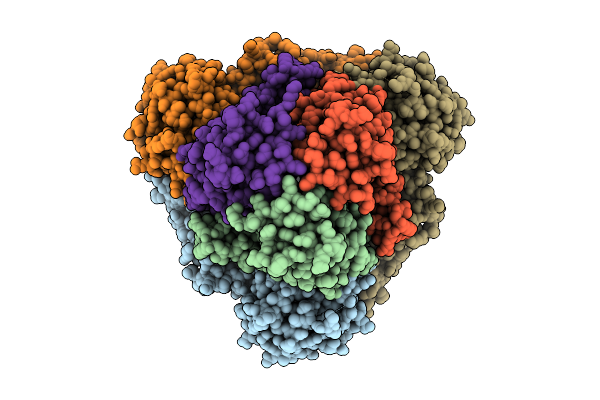 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: FE2, FES, HCI |
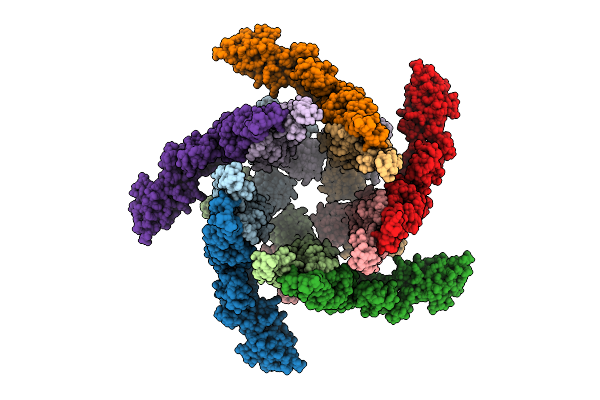 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: STRUCTURAL PROTEIN |
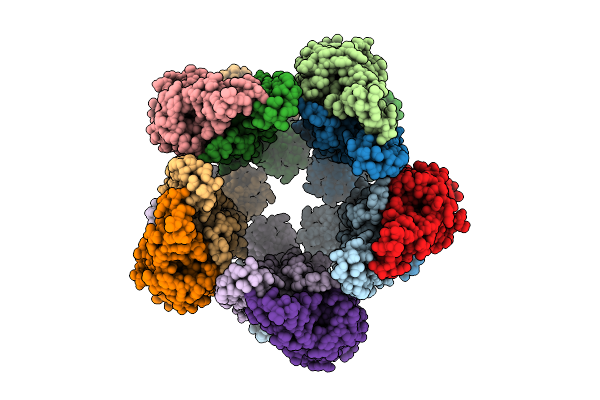 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: STRUCTURAL PROTEIN |
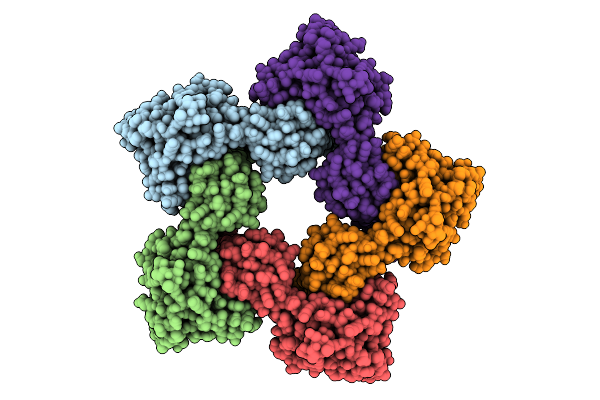 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: STRUCTURAL PROTEIN |
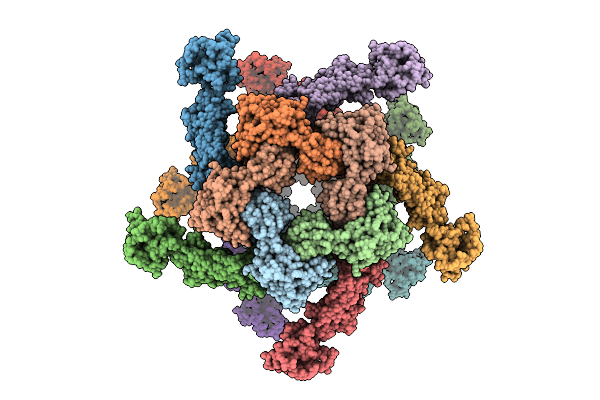 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: STRUCTURAL PROTEIN |
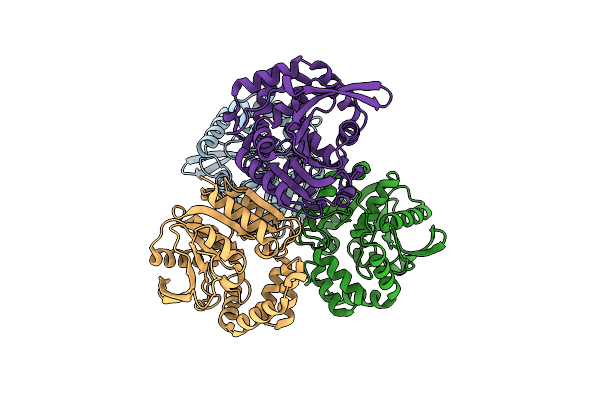 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: HYDROLASE |
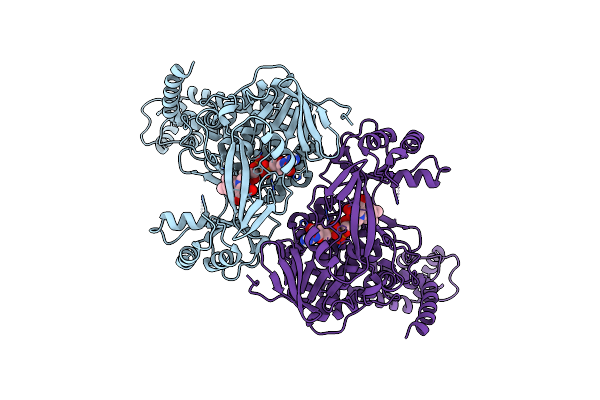 |
Cryoem Structure Of A Trna Uridine 5-Carboxymethylaminomethyl Modification Enzyme Gida
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: HYDROLASE Ligands: FAD |
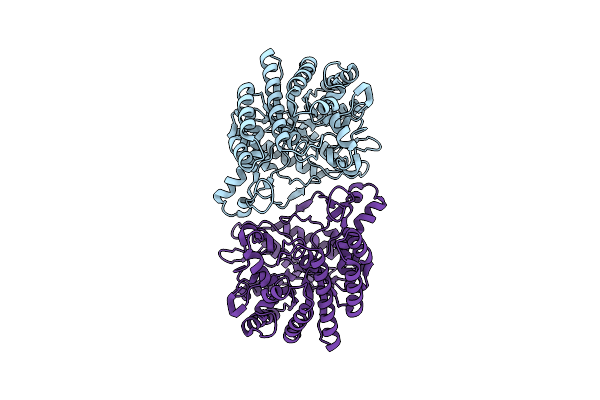 |
Organism: Caldicellulosiruptor owensensis (strain atcc 700167 / dsm 13100 / ol)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: HYDROLASE |
 |
Organism: Acetivibrio thermocellus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: HYDROLASE |
 |
Organism: Parageobacillus caldoxylosilyticus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structure Of A Beta-1,4-Endoglucanase From Bispora Sp. Mey-1 In Complex With Mannotetrose
Organism: Bispora sp. mey-1
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: NAG, PEG |
 |
Cryoem Structure Of A 2,3-Hydroxycinnamic Acid 1,2-Dioxygenase Mhpb In Substrate Bound Form
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: BIOSYNTHETIC PROTEIN Ligands: FE2, A1EG2 |
 |
Cryoem Structure Of The Trifunctional Nad Biosynthesis/Regulator Protein Nadr In Complex With Nad
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: CELL INVASION Ligands: NAD, PO4 |
 |
Organism: Streptomyces hygrospinosus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: TRANSFERASE |
 |
Cryoem Structure Of The Salmonella Effector Inositol Phosphate Phosphatase Sopb
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: LIPID BINDING PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: LIPID BINDING PROTEIN |
 |
Cryoem Structure Of A 2,3-Hydroxycinnamic Acid 1,2-Dioxygenase Mhpb In Apo Form
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: BIOSYNTHETIC PROTEIN |
 |
Cryoem Structure Of 3-Phenylpropionate/Cinnamic Acid Dioxygenase Hcae-Hcaf Complex
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: SOLUTION NMR Release Date: 2023-07-05 Classification: CHAPERONE |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: SOLUTION NMR Release Date: 2023-07-05 Classification: CHAPERONE |