Search Count: 59
 |
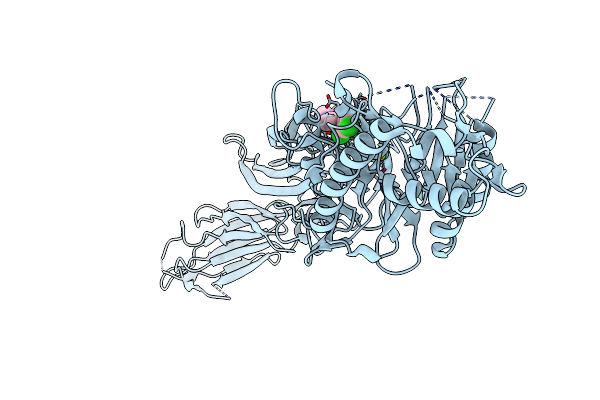
Potent Inhibition Of The Protein Arginine Deiminases (Pad1-4) By Targeting A Ca2+ Dependent Allosteric Binding Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: CA, A1A8O |
 |
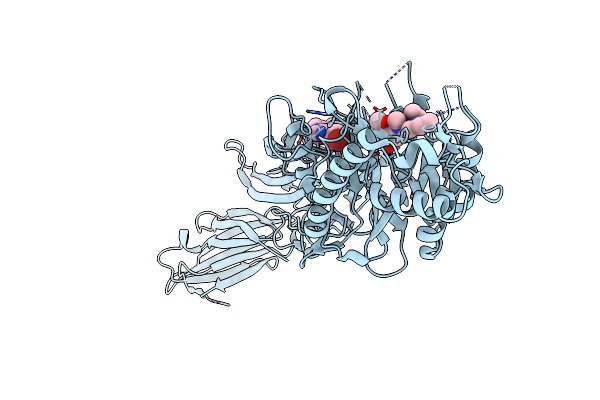
Inhibiting Peptidylarginine Deiminases (Pad1-4) By Targeting A Ca2+ Dependent Allosteric Binding Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2025-05-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, A1BZK |
 |
Potent Inhibition Of The Protein Arginine Deiminases (Pad1-4) By Targeting A Ca2+ Dependent Allosteric Binding Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2025-05-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: A1A9P, 3YZ |
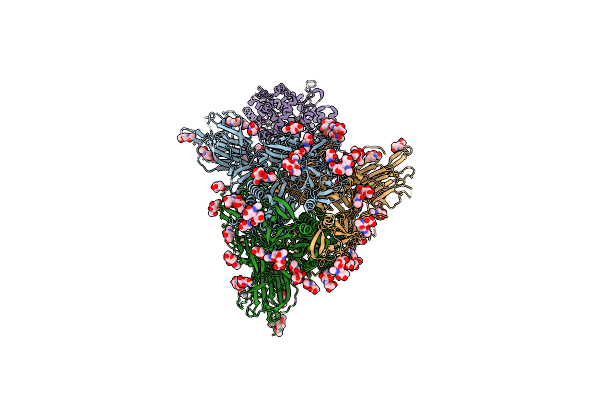 |
Cryo-Em Structure Of Sars-Cov-2 D614G S With One Ace2 Receptor Binding (Rb1) In Prefusion Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
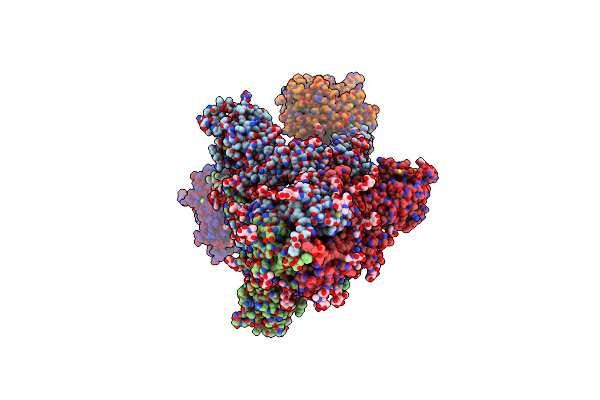 |
Cryo-Em Structure Of Sars-Cov-2 D614G S With Two Ace2 Receptors Binding (Rb2) In Prefusion Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 D614G S With Three Ace2 Receptors Binding (Rb3) In Prefusion Conformation (Focused Refinement Of Ntd-Sd1-Rbd-Ace2)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 D614G S With Three Ace2 Receptors Binding (Rb3) In Prefusion Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 S Trimer In The Early Fusion Intermediate Conformation (E-Fic) (Focused Refinement Of Ntd-Sd1-Rbd-Ace2)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
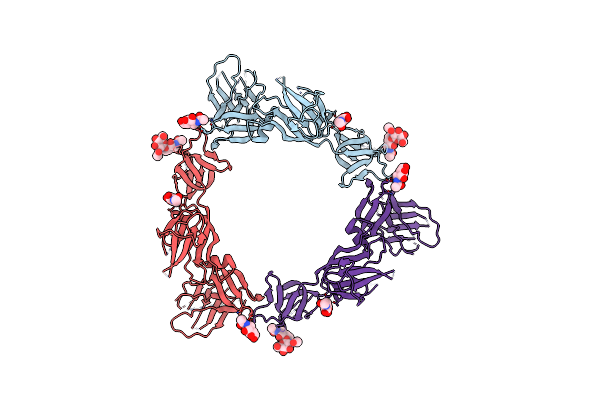
Cryo-Em Structure Of Sars-Cov-2 S Trimer In The Early Fusion Intermediate Conformation (E-Fic) (Focused Refinement Of Intact S2)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
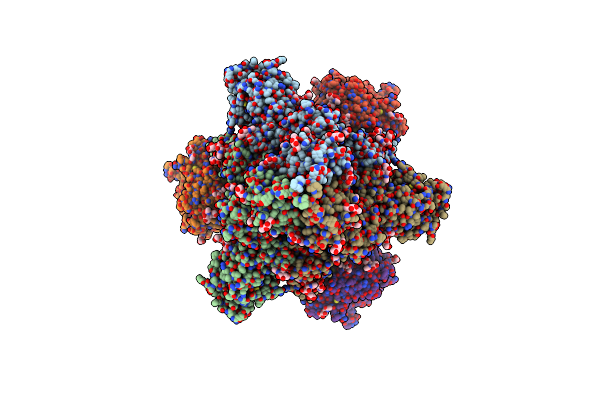
Cryo-Em Structure Of Sars-Cov-2 S Trimer In The Early Fusion Intermediate Conformation (E-Fic) (Focused Refinement Of S-Bottom)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
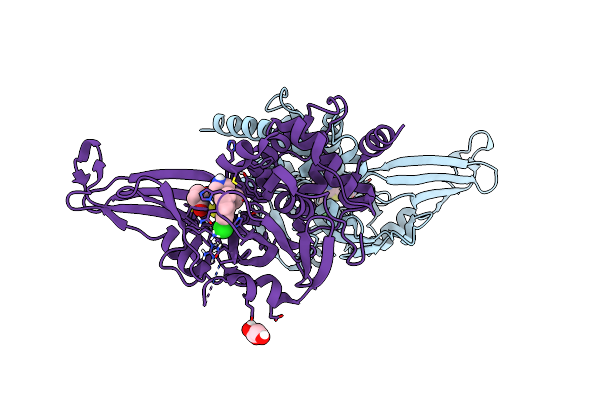
Cryo-Em Structure Of Sars-Cov-2 S Trimer In The Early Fusion Intermediate Conformation (E-Fic)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: VIRAL PROTEIN/HTDROLASE Ligands: NAG |
 |
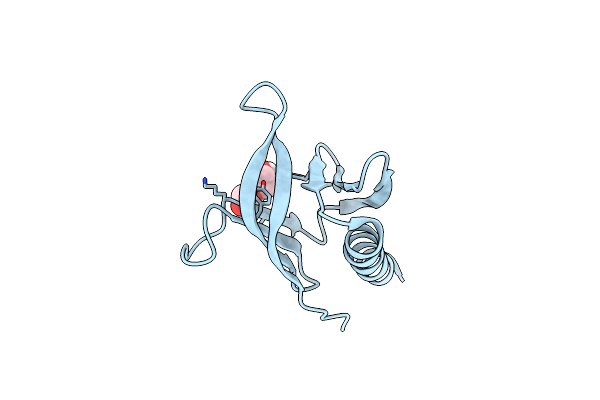
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2023-09-20 Classification: HYDROLASE/INHIBITOR Ligands: QBL, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-05-03 Classification: CYTOSOLIC PROTEIN Ligands: PEG |
 |
Crystal Structure Of N-Terminal Ph Domain Of Arap3 Protein In Complex With Inositol 1,3,4,5-Tetrakisphosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2023-05-03 Classification: CYTOSOLIC PROTEIN Ligands: 4PT |
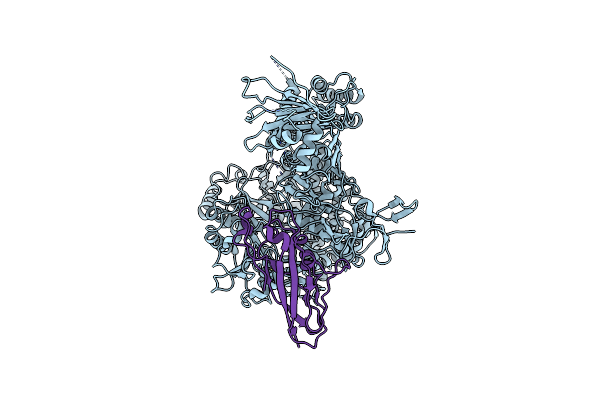 |
Structural Basis For Cspg4 As A Receptor For Tcdb And A Therapeutic Target In Clostridioides Difficile Infection
Organism: Clostridioides difficile, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-06-09 Classification: TOXIN Ligands: ZN |
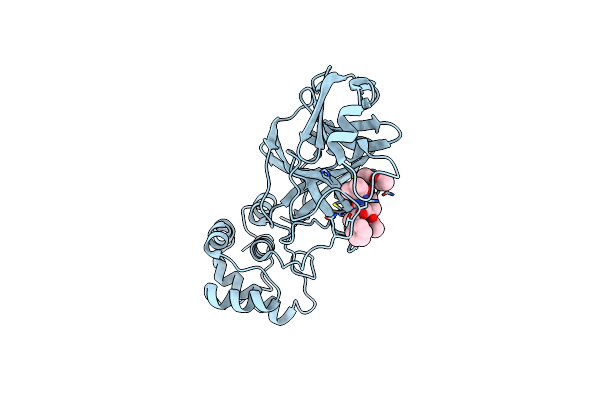 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-03-17 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: TG3 |
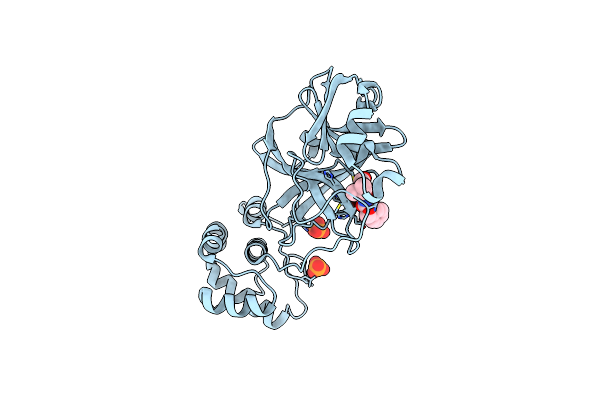 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2021-03-10 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: UED, PO4 |
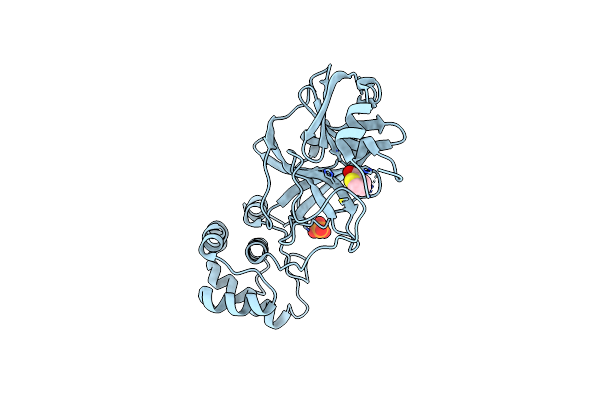 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2021-03-10 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: LW1, PO4 |
 |
Crystal Structure Of Sars-Cov-2 3Cl Protease In Complex With Compound 4 In Space Group P1
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-03-10 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: TG3, BTB |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-03-03 Classification: VIRAL PROTEIN, HYDROLASE Ligands: PO4 |

