Search Count: 62
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: DE NOVO PROTEIN Ligands: 308 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: DE NOVO PROTEIN Ligands: UNX, 308 |
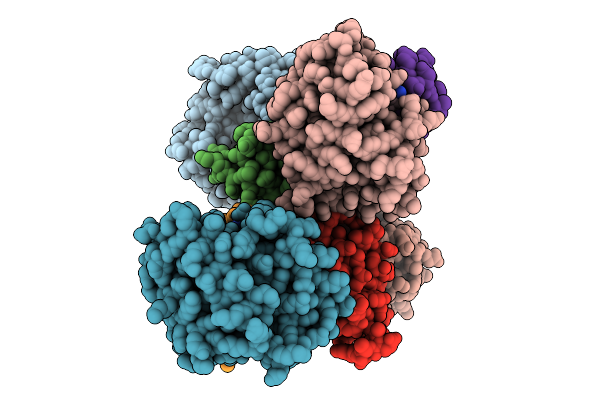 |
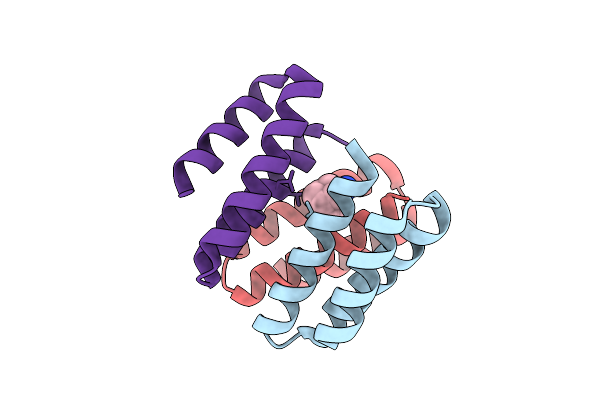
Crystal Structure Of De Novo Designed Amantadine Induced Heterodimer Daid23.4
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: DE NOVO PROTEIN Ligands: 308 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: DE NOVO PROTEIN Ligands: 308 |
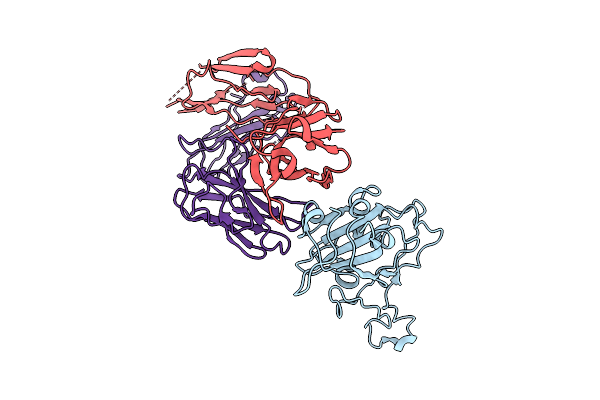 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
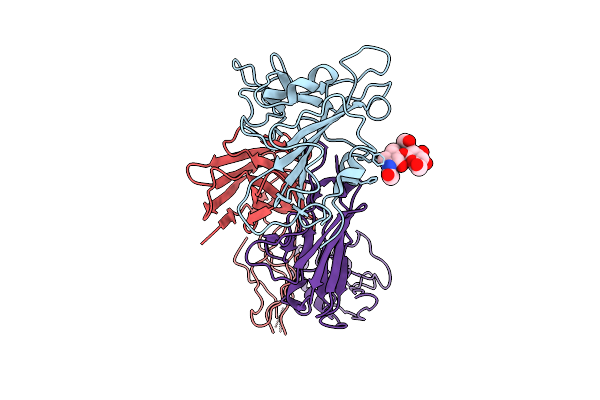 |
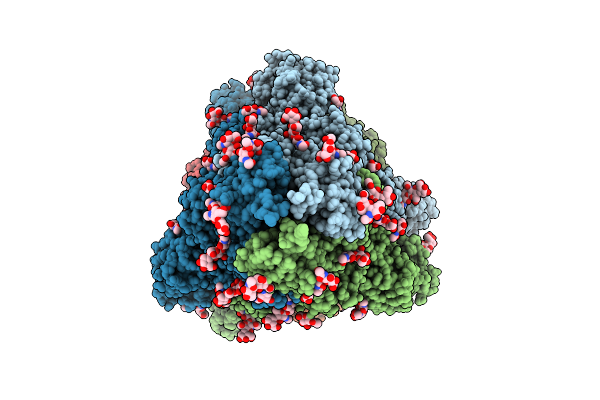
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-183
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
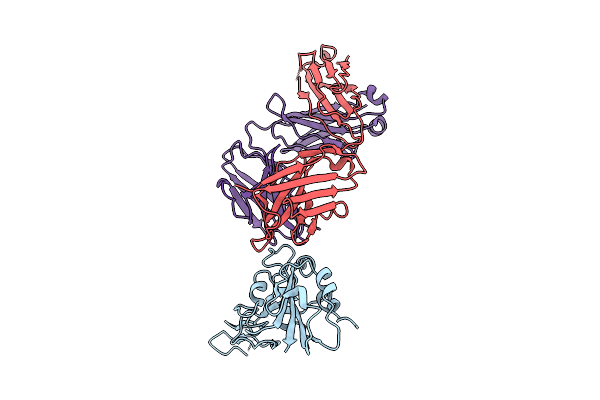
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-171
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
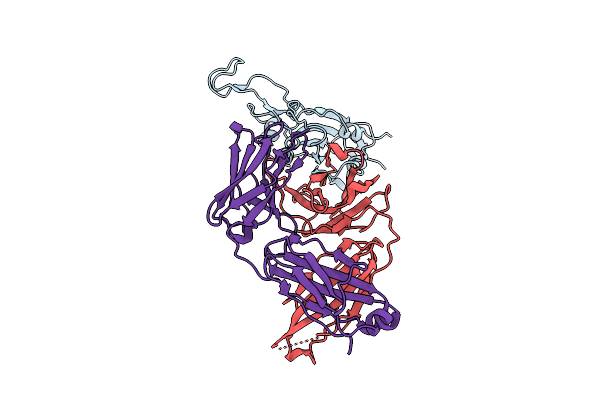 |
The Local Refined Map Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
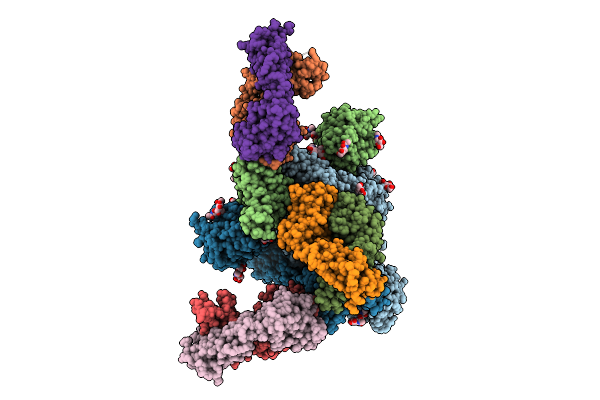 |
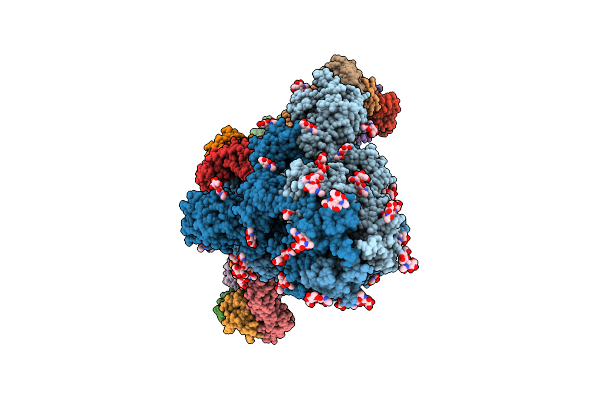
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-203
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Structure Of Sars-Cov-2 Eg.5.1 Variant Spike Protein Complexed With Antibody Xgi-198
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C2-5 (Lro-C2-5)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C2-35 (Lro-C2-35) At Acidic Ph (Ph 4.5)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C2-35 (Lro-C2-35) At Basic Ph (Ph 8.5)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C3-7 (Lro-C3-7)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C4-13 (Lro-C4-13)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Oligomer C5-1 (Lro-C5-1)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Heterodimer 2 (Lrd-2)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of De Novo Designed Light-Responsive Heterodimer 7 (Lrd-7)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DE NOVO PROTEIN |
 |
Organism: Populus trichocarpa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-03-12 Classification: METAL BINDING PROTEIN Ligands: NI |

