Search Count: 33
 |
Organism: Human gammaherpesvirus 8
Method: ELECTRON MICROSCOPY Release Date: 2025-01-08 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-06-19 Classification: TRANSFERASE/PROTEIN BINDING Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-05-29 Classification: DNA BINDING PROTEIN Ligands: 8JA |
 |
Organism: Brassica oleracea
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
A Cryo-Em Structure Of B. Oleracea Rna Polymerase V Elongation Complex At 2.73 Angstrom
Organism: Brassica oleracea
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryoem Structure Of Arabidopsis Ros1 In Complex With Tg Mismatch Dsdna At 3.3 Angstroms Resolution
Organism: Homo sapiens, Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: DNA BINDING PROTEIN/DNA Ligands: SF4 |
 |
Cryoem Structure Of Arabidopsis Ros1 In Complex With 5Mc-Dsdna At 3.1 Angstroms Resolution
Organism: Homo sapiens, Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: DNA BINDING PROTEIN/DNA Ligands: SF4 |
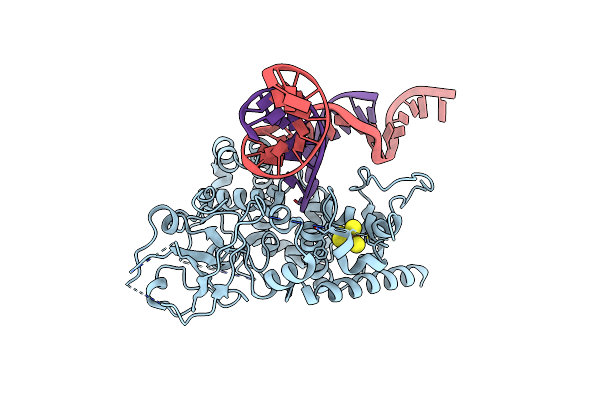 |
Cryoem Structure Of Arabidopsis Ros1 In Complex With A Covalent-Linked Reaction Intermediate At 3.9 Angstroms Resolution
Organism: Homo sapiens, Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2022-11-30 Classification: DNA BINDING PROTEIN/DNA Ligands: SF4 |
 |
Crystal Structure Of Ttnm, A Fe(Ii)-Alpha-Ketoglutarate-Dependent Hydroxylase From The Tautomycetin Biosynthesis Pathway In Streptomyces Griseochromogenes At 2 A.
Organism: Streptomyces griseochromogenes
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-07-06 Classification: OXIDOREDUCTASE Ligands: FE2, CL |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-06-08 Classification: PLANT PROTEIN |
 |
Organism: Zea mays
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: PLANT PROTEIN |
 |
Organism: Zea mays
Method: ELECTRON MICROSCOPY Release Date: 2022-06-08 Classification: PLANT PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-09-15 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-15 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-09-15 Classification: DNA BINDING PROTEIN/DNA |
 |
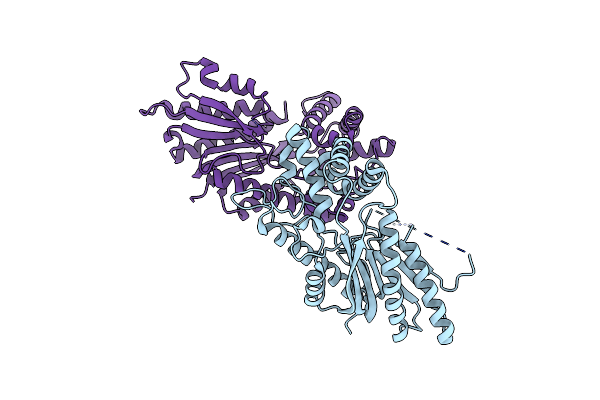
Crystal Structure Of Tea N9-Methyltransferase Cktcs In Complex With Sah And 1,3,7-Trimethyluric Acid
Organism: Camellia sinensis var. assamica
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2020-03-04 Classification: TRANSFERASE Ligands: SAH, EXU |
 |
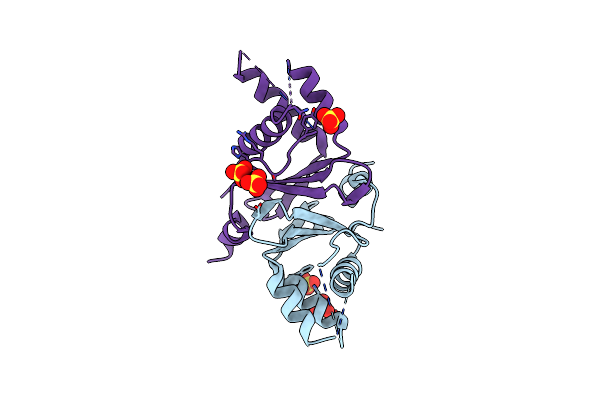
Crystal Structure Of A N-Methyltransferase Cktbs From Camellia Assamica Var. Kucha
Organism: Camellia sinensis var. assamica
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2020-03-04 Classification: TRANSFERASE |
 |
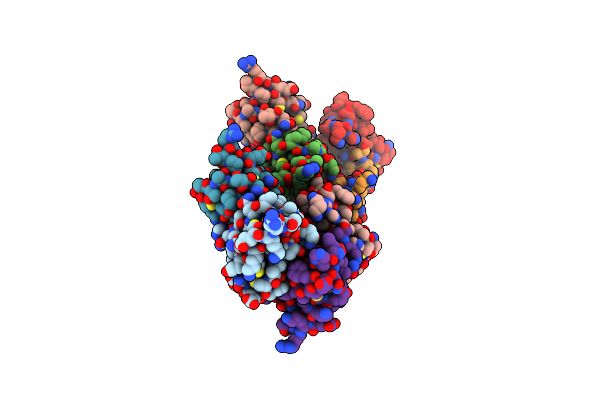
Organism: Schizosaccharomyces pombe 972h-
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-02-13 Classification: RNA BINDING PROTEIN Ligands: SO4 |
 |
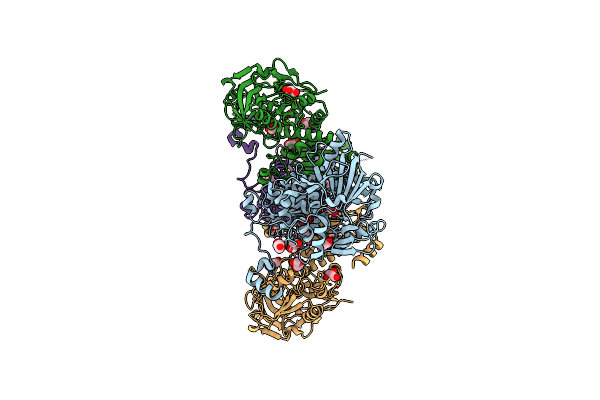
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-10-24 Classification: LIGASE Ligands: ZN |
 |
Crystal Structure Of The Ttnd Decarboxylase From The Tautomycetin Biosynthesis Pathway Of Streptomyces Griseochromogenes, Apo Form At 2.6 A Resolution (P212121)
Organism: Streptomyces griseochromogenes
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2018-10-03 Classification: LYASE Ligands: MG, GOL, UNL |

