Search Count: 64
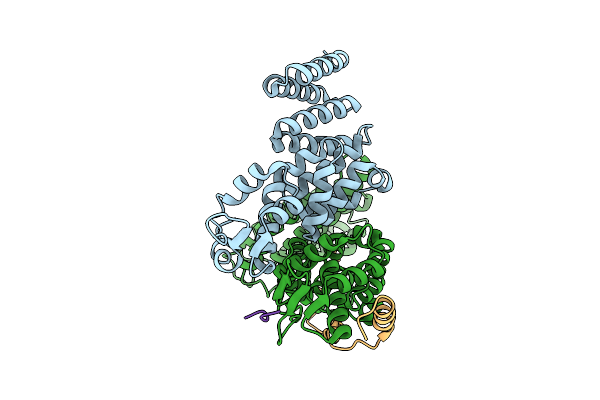 |
Cryo-Em Structure Of The Human Trex-2.1 Complex (Leng8/Pcid2/Dss1) Bound To The N-Terminal Motif Of Ddx39B(Uap56)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: RNA BINDING PROTEIN/Hydrolase |
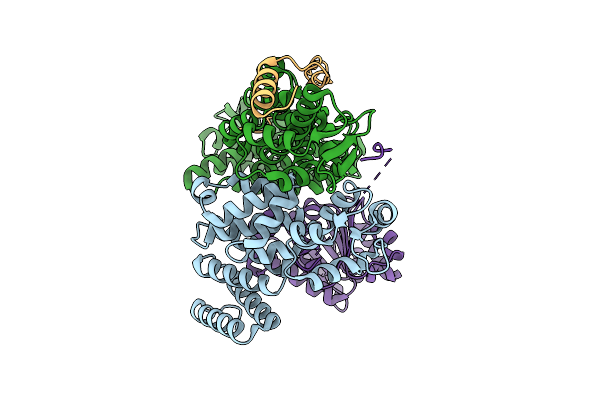 |
Cryo-Em Structure Of The Human Trex-2.1 Complex (Leng8/Pcid2/Dss1) Bound To Ddx39B(Uap56)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: RNA BINDING PROTEIN/Hydrolase |
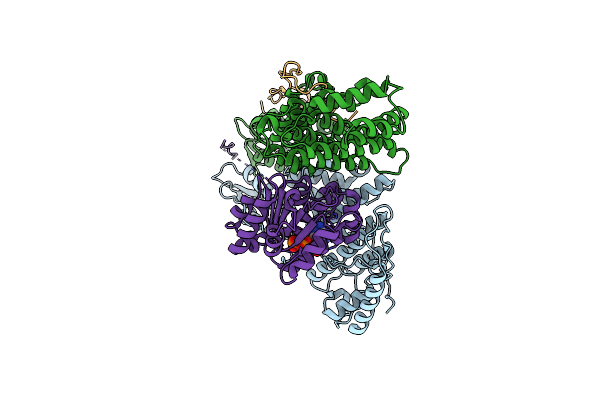 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: RNA BINDING PROTEIN Ligands: ADP, MG |
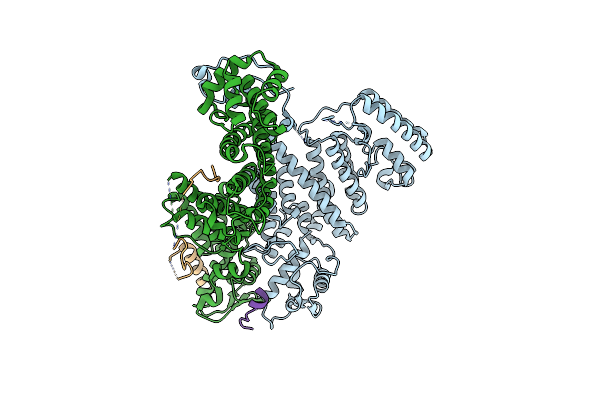 |
Crystal Structure Of The Trex-2 Complex In Complex With The N-Terminal Motif Of Sub2
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Release Date: 2025-03-19 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of Irak4-Hsa Complexed With N-[(2R)-2-Fluoro-3-Hydroxy-3-Methylbutyl]-6-[(5-Fluoropyri-Yl)Amino]-4-[(Propan-2-Yl)Amino]Pyridine-3-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-02-12 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZVD, SO4 |
 |
Crystal Structure Of Irak4-Hsa Complexed With Bms-986126; 6-((5-Cyano-2-Pyrimidinyl)Amino)-N-((2R)-2-Fluoro-3-Hydroxhylbutyl)-4-(Isopropylamino)Nicotinamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2025-02-12 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZVG, SO4 |
 |
Crystal Structure Of Irak4-Hsa Complexed With Bms-986147; 6-{5-Cyano-1H-Pyrazolo[3,4-B]Pyridin-1-Yl}-N-[(2R)-2-Fluorroxy-3-Methylbutyl]-4-[(Propan-2-Yl)Amino]Pyridine-3-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2025-02-12 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: SO4, ZVA |
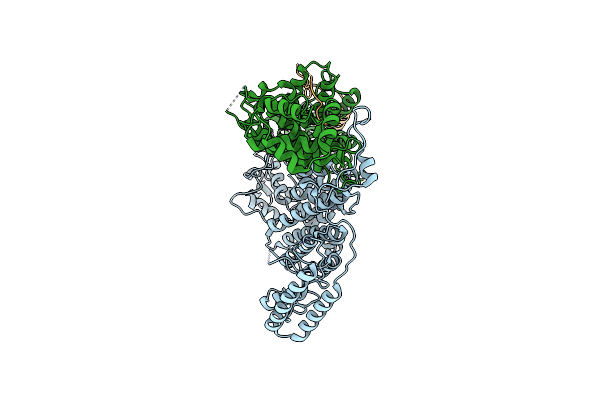 |
Cryo-Em Structure Of The Trex-2 Complex In Complex With The N-Terminal Motif Of Sub2
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: RNA BINDING PROTEIN |
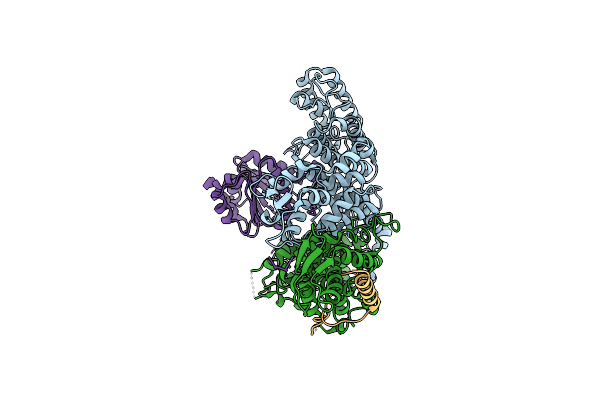 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: RNA BINDING PROTEIN |
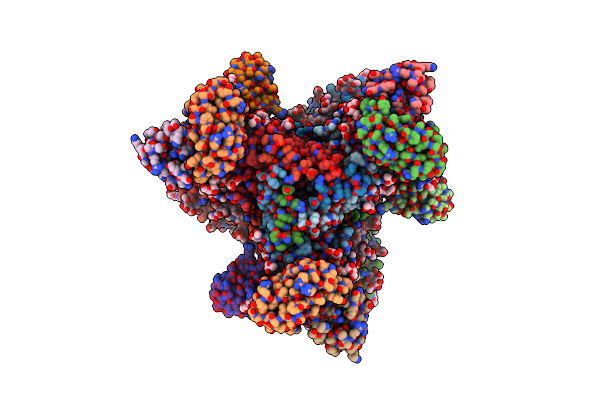 |
Structure Of Vrc44.01 Fab In Complex With 3Bnc117-Purified C1080.C3 Rns Sosip.664 Hiv-1 Env Trimer
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, PO4 |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: TRANSPORT PROTEIN Ligands: NAG, Y7H |
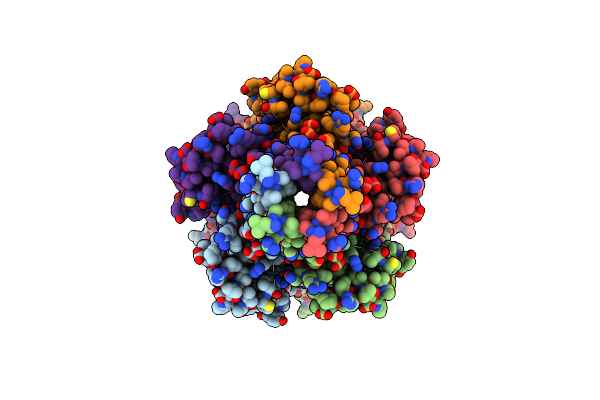 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: TRANSPORT PROTEIN Ligands: Y82, NAG |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: TRANSPORT PROTEIN Ligands: SRO, NAG |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: TRANSPORT PROTEIN Ligands: NAG, SRO |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: TRANSPORT PROTEIN Ligands: NAG, Y82 |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-12-27 Classification: TRANSPORT PROTEIN Ligands: Y7H, NAG |
 |
Organism: Severe fever with thrombocytopenia syndrome virus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: VIRAL PROTEIN |
 |
Organism: Severe fever with thrombocytopenia syndrome virus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: VIRAL PROTEIN |
 |
Organism: Severe fever with thrombocytopenia syndrome virus
Method: ELECTRON MICROSCOPY Release Date: 2023-07-19 Classification: VIRAL PROTEIN |
 |
Organism: Striga hermonthica
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2022-01-05 Classification: HYDROLASE |

