Search Count: 296
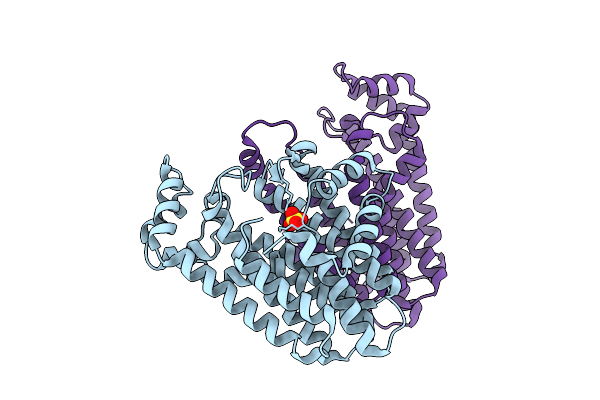 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: SO4 |
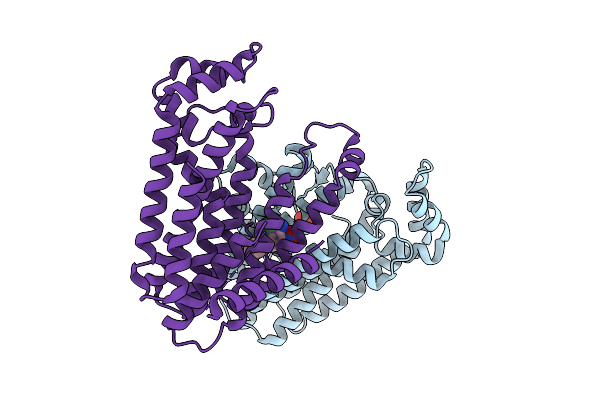 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: SO4, A1L38 |
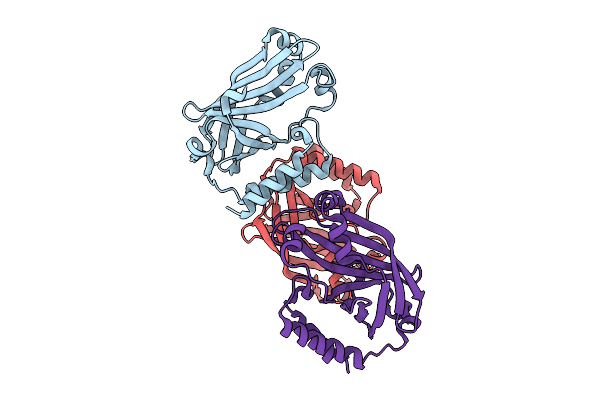 |
Organism: Oryza sativa subsp. japonica
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: UNKNOWN FUNCTION |
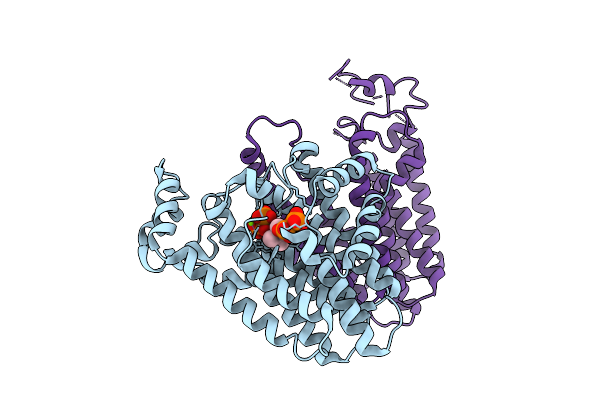 |
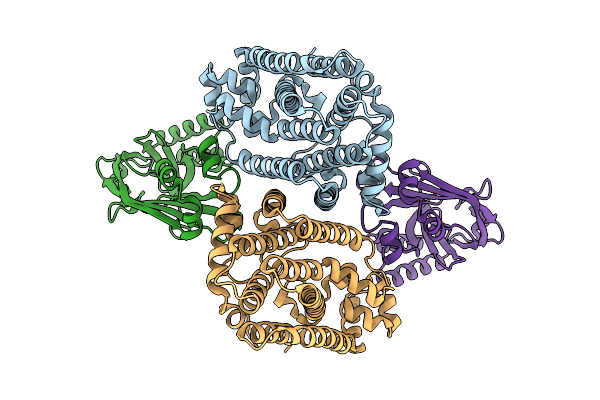
Crystal Structure Of Ossps3 Complexed With Zoledronate And Isopentenyl Diphosphate
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: IPE, ZOL, MG |
 |
Organism: Oryza sativa subsp. japonica
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSFERASE |
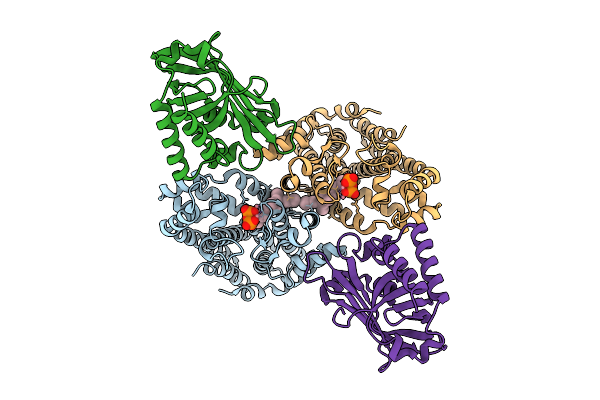 |
Cryo-Em Structure Of The Sps3-Fbn5 Complex From Oryza Sativa In Complex With Cobalt And Geranylgeranyl S-Thiodiphosphate (Ggspp)
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: GGS, CO |
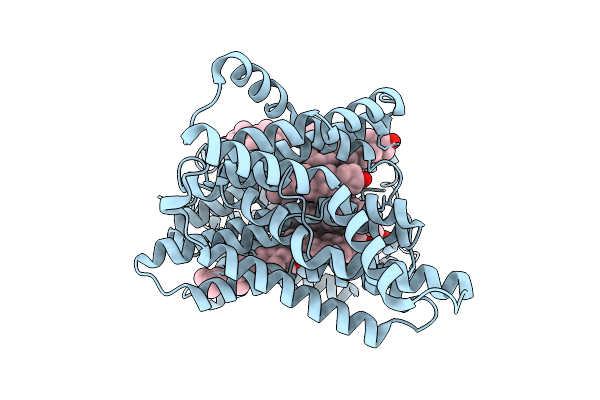 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: CLR |
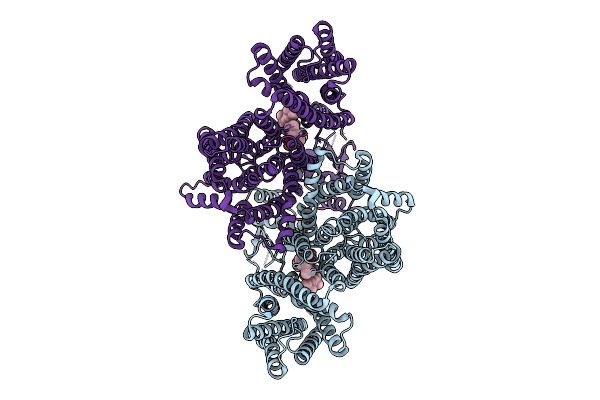 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: Y01, CLR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Map Of Msmeg_3496 In Complex With Acpm, Size Exclusion Chromatography Peak1
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Map Of Msmeg_3496 In Complex With Acpm, Size Exclusion Chromatography Peak2
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv), Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Wuhan spiny eel influenza virus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Escherichia phage
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
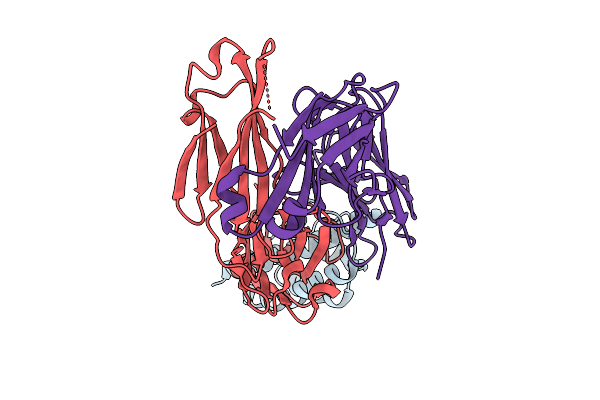 |
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: CYTOKINE/IMMUNE SYSTEM |
 |
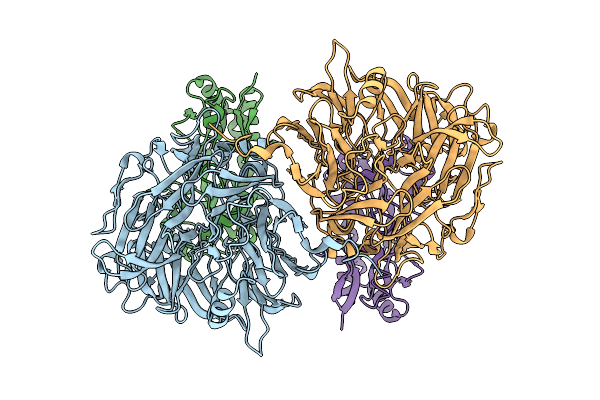
Structural Insights Into Il-31 Signaling Inhibition By A Neutralization Antibody
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: CYTOKINE/IMMUNE SYSTEM |
 |
Organism: Methylorubrum extorquens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: PROTEIN BINDING |

