Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
X-Ray Structure Of The Gdp-6-Deoxy-4-Keto-D-Lyxo-Heptose-4-Reductase From Campylobacter Jejuni Hs:15
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-11-09 Classification: BIOSYNTHETIC PROTEIN, OXIDOREDUCTASE Ligands: GDP, NDP, EDO |
 |
X-Ray Crystal Structure Of Gdp-D-Glycero-D-Manno-Heptose 4,6-Dehydratase From Campylobacter Jejuni
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-07-06 Classification: OXIDOREDUCTASE Ligands: NAD, GDP, EDO |
 |
Crystal Structure Of The Gdp-D-Glycero-4-Keto-D-Lyxo-Heptose-3-Epimerase From Campylobacter Jejuni, Serotype Hs:23/36
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-06-29 Classification: ISOMERASE Ligands: GDP, K, EDO, NA |
 |
Crystal Structure Of The Gdp-D-Glycero-4-Keto-D-Lyxo-Heptose-3,5-Epimerase From Campylobacter Jejuni, Serotype Hs:42
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-06-29 Classification: ISOMERASE Ligands: GDP, CL, EDO |
 |
Crystal Structure Of The Gdp-D-Glycero-4-Keto-D-Lyxo-Heptose-3-Epimerase From Campylobacter Jejuni, Serotype Hs:3
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-06-22 Classification: ISOMERASE Ligands: GDP, CL, TMA |
 |
Crystal Structure Of The Gdp-D-Glycero-4-Keto-D-Lyxo-Heptose-3,5-Epimerase From Campylobacter Jejuni, Serotype Hs:15
Organism: Campylobacter jejuni
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-06-22 Classification: ISOMERASE Ligands: GDP, CL |
 |
Organism: Sphingobium sp. tcm1
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2016-07-13 Classification: HYDROLASE Ligands: MN |
 |
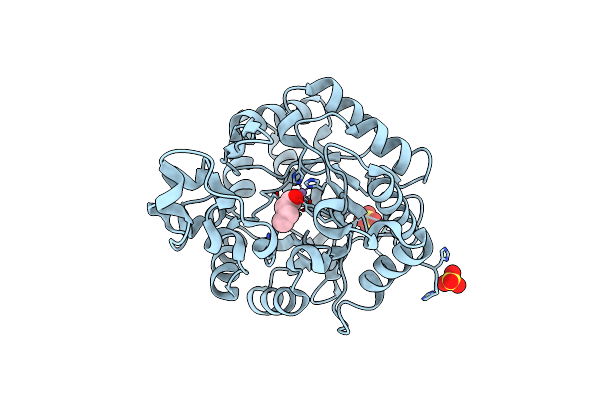
Crystal Structure Of The Sphingobium Sp. Tcm1 Phosphotriesterase Without The Binuclear Manganese Center
Organism: Sphingobium sp. tcm1
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-07-13 Classification: HYDROLASE |
 |
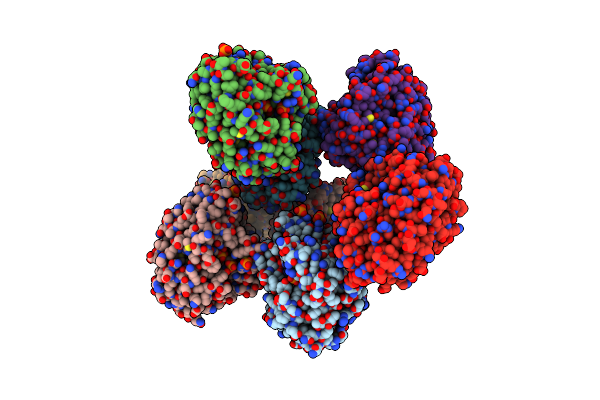
Crystal Structure Of Amidohydrolase Pmi1525 (Target Efi-500319) From Proteus Mirabilis Hi4320, A Complex With Butyric Acid And Manganese
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-07-30 Classification: HYDROLASE Ligands: MN, BUA, SO4 |
 |
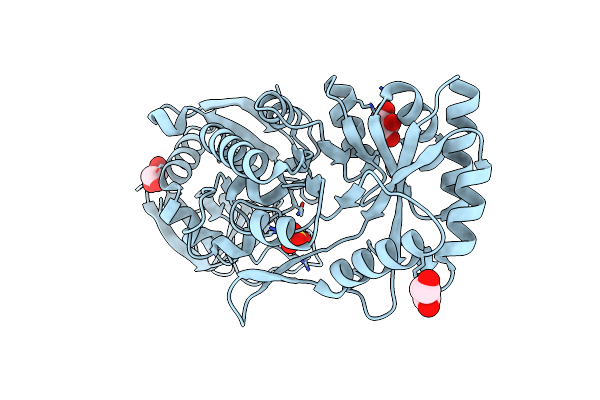
Crystal Structure Of Phosphotriesterase Homology Protein From Escherichia Coli Complexed With Phosphate In Active Site
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2014-06-25 Classification: HYDROLASE Ligands: ZN, PO4, BGC |
 |
Crystal Structure Of The Pyridoxal-5'-Phosphate Dependent Protein Yhfx From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-05-21 Classification: UNKNOWN FUNCTION Ligands: SO4, GOL, CL |
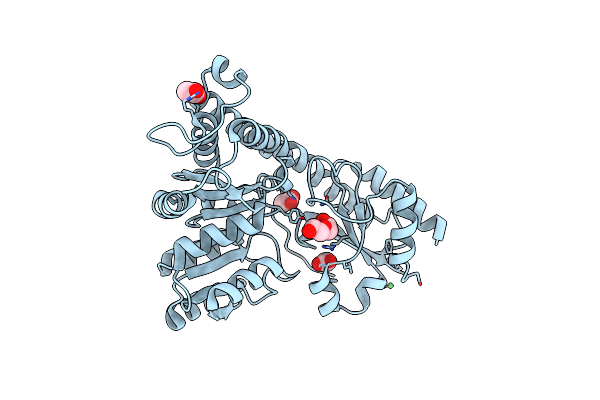 |
Crystal Structure Of The Pyridoxal-5'-Phosphate Dependent Protein Yhfs From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-03-05 Classification: UNKNOWN FUNCTION Ligands: NI, ACY, PEG |
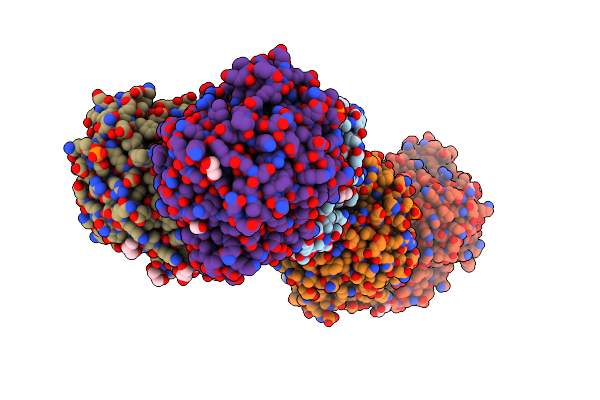 |
Organism: Listeria monocytogenes serotype 4b str. h7858
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-11-23 Classification: HYDROLASE Ligands: GOL, ZN, PO4 |
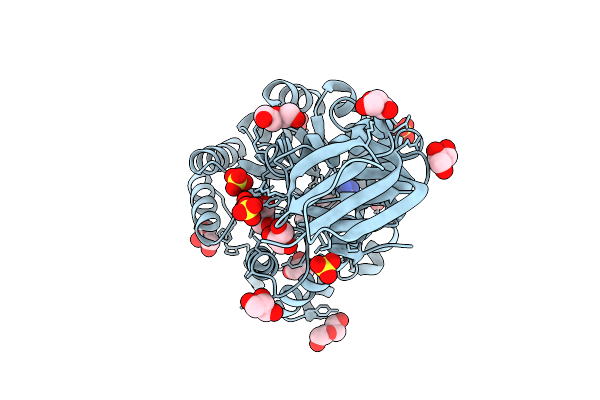 |
Crystal Structure Of L-Lysine, L-Arginine Carboxypeptidase Cc2672 From Caulobacter Crescentus Cb15 Complexed With N-Methyl Phosphonate Derivative Of L-Arginine
Organism: Caulobacter vibrioides
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-07-28 Classification: HYDROLASE Ligands: ZN, M3R, SO4, GOL, PEG |
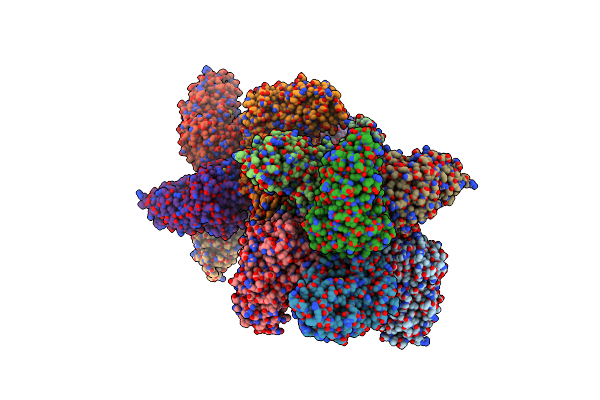 |
Crystal Structure Of Prolidase Eah89906 Complexed With N-Methylphosphonate-L-Proline
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2010-06-02 Classification: HYDROLASE Ligands: ZN, LWY |
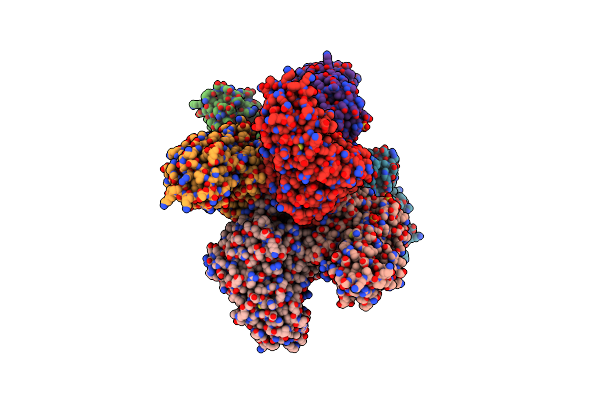 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-04-28 Classification: HYDROLASE Ligands: ZN, SO4, GOL, CO3 |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-06-30 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2008-12-16 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: ZN |
 |
Crystal Structure Of Zn-Dependent Arginine Carboxypeptidase Complexed With Zinc
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2008-08-05 Classification: HYDROLASE Ligands: ZN, ARG, GOL |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-11-27 Classification: HYDROLASE Ligands: ARG, GOL, MG |