Search Count: 17
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-12-05 Classification: OXIDOREDUCTASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-09-05 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of N-Terminal 57 Residue Deletion Mutant Of E. Coli Ccmg Protein(Residues 58-185)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-09-05 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-03-15 Classification: ISOMERASE |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-02-01 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2005-02-01 Classification: ELECTRON TRANSPORT Ligands: HEM |
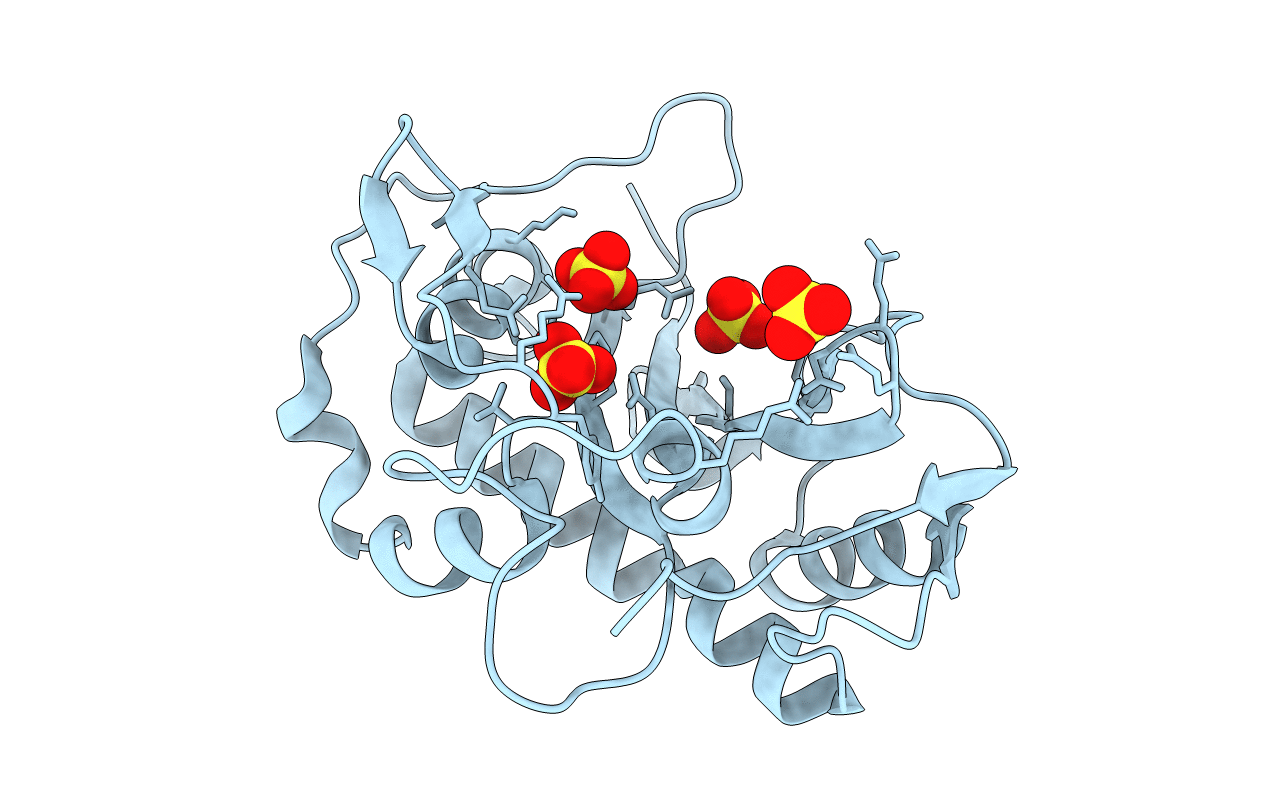 |
The Three-Dimensional Structure And X-Ray Sequence Reveal That Trichomaglin Is A Novel S-Like Ribonuclease
Organism: Trichosanthes lepiniana
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-06-22 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of F35Y Mutant Of Trypsin-Solubilized Fragment Of Cytochrome B5
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-09-11 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-09-04 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2002-09-04 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
Catalytic Mechanism Of Quinoprotein Methanol Dehydrogenase: A Theoretical And X-Ray Crystallographic Investigation
Organism: Methylophilus methylotrophus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2001-01-24 Classification: OXIDOREDUCTASE Ligands: CA, PQQ |
 |
Crystal Structure Of Recombinant Trypsin-Solubilized Fragment Of Cytochrome B5
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2000-08-09 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
Crystal Structure Of Val61His Mutant Of Trypsin-Solubilized Fragment Of Cytochrome B5
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-08-09 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
Crystal Structure Of The Complex Of Trichosanthin With Adenine, Obtained From Trichosanthin Complexed With The Dinucleotide Apg
Organism: Trichosanthes kirilowii
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2000-04-24 Classification: HYDROLASE Ligands: ADE |
 |
Organism: Momordica charantia
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 1999-06-07 Classification: RIBOSOME-INACTIVATING PROTEIN |
 |
Crystal Structure Of Trichosanthin-Nadph Complex At 1.7 Angstroms Resolution Reveals Active-Site Architecture
Organism: Trichosanthes kirilowii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1995-07-10 Classification: PROTEIN SYNTHESIS INHIBITOR Ligands: NDP |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1991-01-15 Classification: OXIDOREDUCTASE (CH-OH(D)-CYTOCHROME(A)) Ligands: HEM, FMN, PYR |

