Search Count: 94
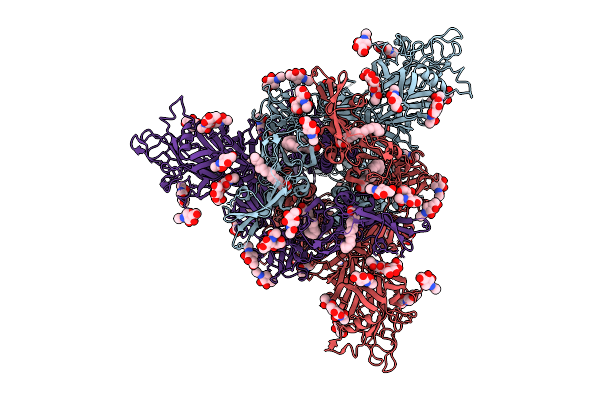 |
Organism: Bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: PLM, NAG, OLA |
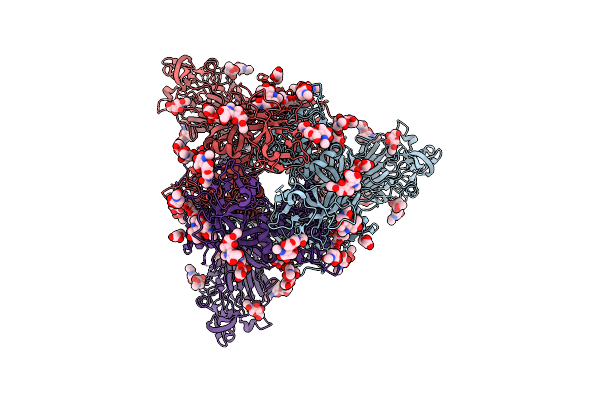 |
Organism: Bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: PLM, NAG |
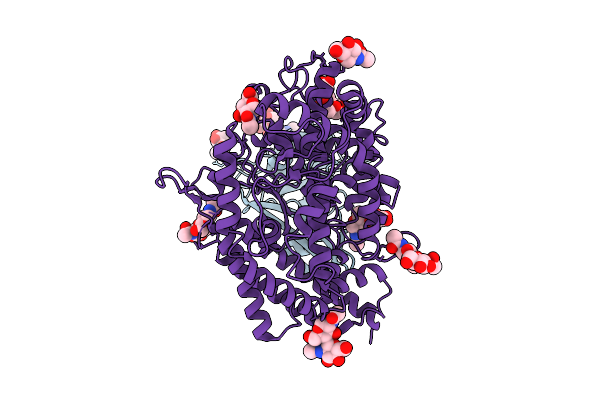 |
Organism: Bat coronavirus hku5, Pipistrellus abramus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: NAG |
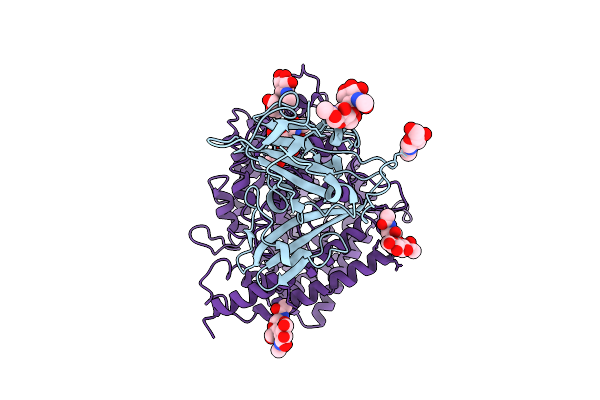 |
Organism: Bat coronavirus hku5, Pitta sordida
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: NAG |
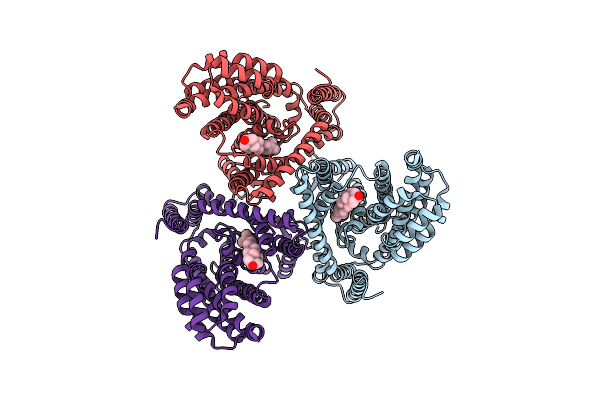 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: TRANSPORT PROTEIN Ligands: CLR |
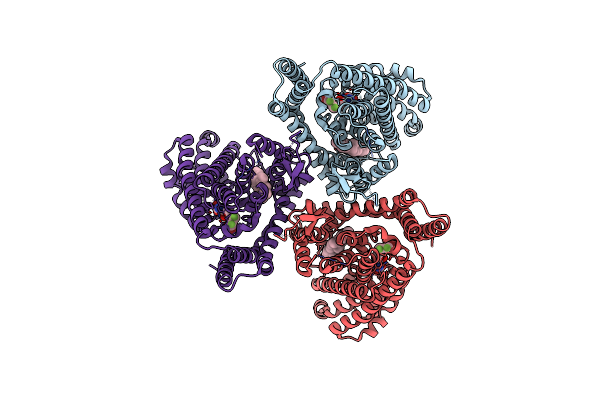 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: TRANSPORT PROTEIN Ligands: CLR, GJ0 |
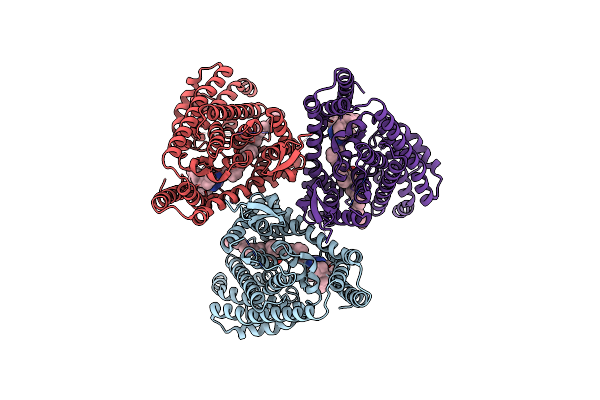 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: TRANSPORT PROTEIN Ligands: CLR, 3PH, A1EDJ |
 |
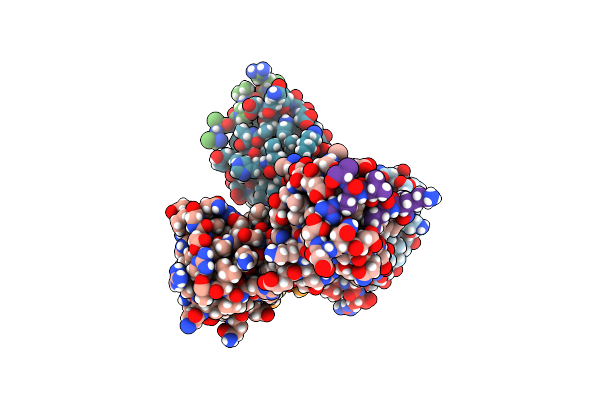
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-03-26 Classification: RIBOSOME Ligands: ZN |
 |
Arf-Gtpase Activating Protein Asap1 Sh3 Domain In Complex With 440 Kd Ankyrin-B Fragment
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: PROTEIN BINDING Ligands: GOL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
The Complex Structure Of The H4B6 Fab With The Rbd Of Omicron Ba.5 S Protein
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of Non-Specific Phospholipase C Replc (Rasamsonia Emersonii)
Organism: Rasamsonia emersonii cbs 393.64
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-07-10 Classification: HYDROLASE Ligands: NAG |
 |
Organism: Human coronavirus hku1 (isolate n2)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human coronavirus hku1 (isolate n2)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human coronavirus hku1 (isolate n2)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human coronavirus hku1 (isolate n2)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human coronavirus hku1 (isolate n2), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Organism: Human coronavirus hku1 (isolate n2), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Organism: Human coronavirus hku1 (isolate n2)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Human coronavirus hku1 (isolate n2)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: NAG |

