Search Count: 105
 |
Organism: Staphylococcus aureus subsp. aureus n315
Method: SOLUTION NMR Release Date: 2025-10-15 Classification: DNA BINDING PROTEIN |
 |
Organism: Staphylococcus aureus subsp. aureus n315, Synthetic construct
Method: SOLUTION NMR Release Date: 2025-10-15 Classification: DNA BINDING PROTEIN |
 |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: SOLUTION NMR Release Date: 2021-11-10 Classification: DNA BINDING PROTEIN |
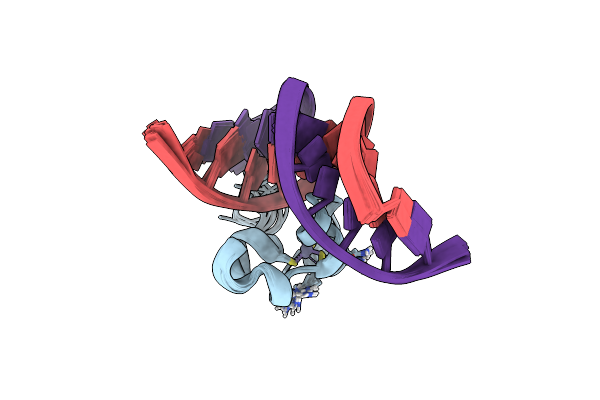 |
Solution Structure Of The C-Clamp Domain From Human Hdbp1 In Complex With Dna
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2021-07-21 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Caulobacter vibrioides (strain na1000 / cb15n)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Caulobacter vibrioides (strain na1000 / cb15n), Caulobacter crescentus na1000
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-08-05 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Caulobacter vibrioides (strain na1000 / cb15n)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2020-06-03 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
2.8 Angstrom Crystal Structure Of Succinate-Acetate Permease From Citrobacter Koseri
Organism: Citrobacter koseri atcc baa-895
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-11-21 Classification: TRANSPORT PROTEIN Ligands: ACT, 78M |
 |
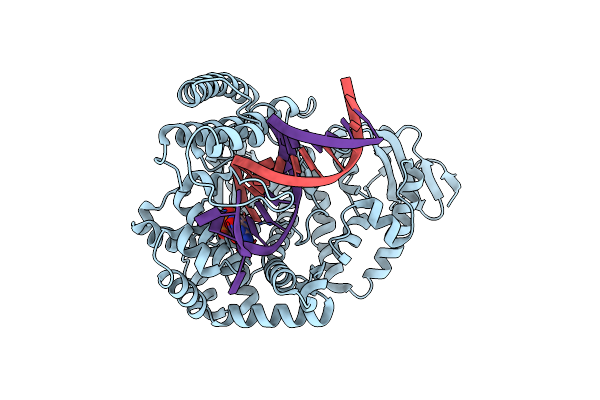
Large Fragment Of Dna Polymerase I From Thermus Aquaticus In A Closed Ternary Complex With The Unnatural Base M-Fc Pair With Datp In The Active Site
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2018-11-21 Classification: TRANSFERASE/DNA Ligands: DTP, MG |
 |
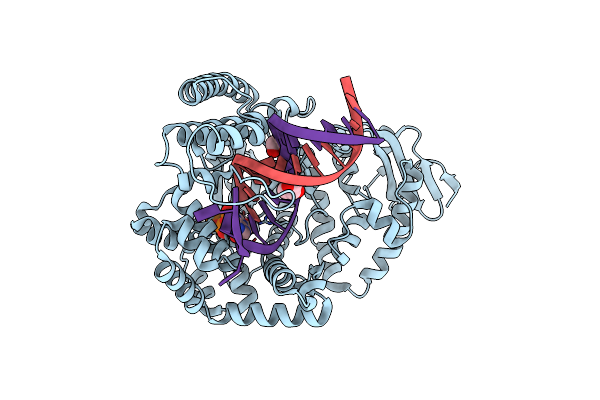
Large Fragment Of Dna Polymerase I From Thermus Aquaticus In A Closed Ternary Complex With The Natural Base Pair 5Fc:Dgtp
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-11-21 Classification: REPLICATION/DNA Ligands: DGT, MG |
 |
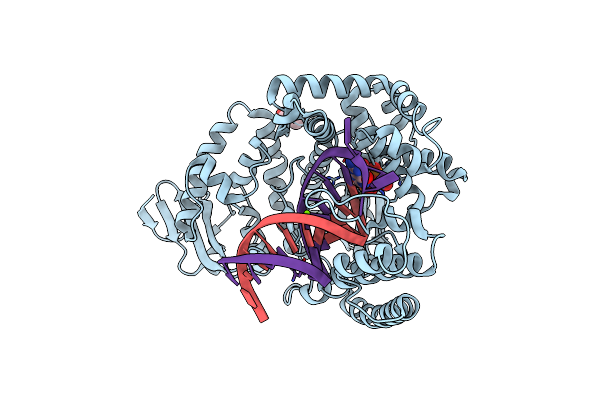
Large Fragment Of Dna Polymerase I From Thermus Aquaticus In A Closed Ternary Complex With With Natural Dt:Datp Base Pair
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2018-11-21 Classification: REPLICATION/DNA Ligands: DTP, MG, GOL |
 |
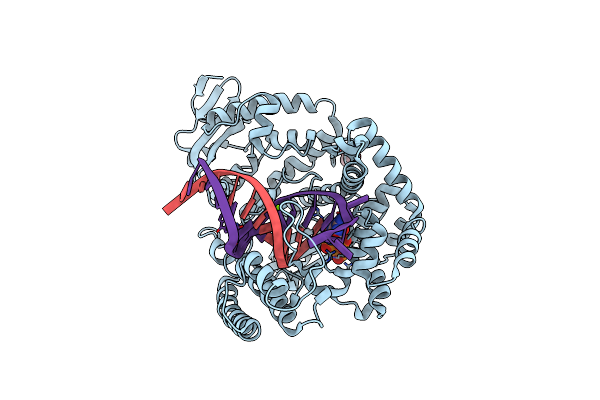
Structure Of Large Fragment Of Dna Polymerase I From Thermus Aquaticus Host-Guest Complex With The Unnatural Base M-Fc Pair With Da
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-11-21 Classification: TRANSFERASE/DNA Ligands: MG, DGT, GOL |
 |
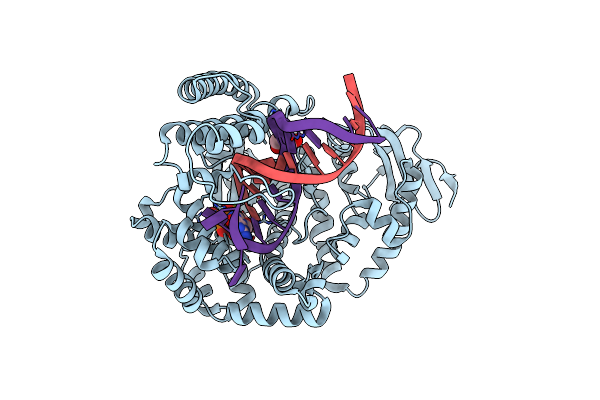
Structure Of Large Fragment Of Dna Polymerase I From Thermus Aquaticus Host-Guest Complex With The Unnatural Base I-Fc Pair With Da
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2018-11-21 Classification: TRANSFERASE/DNA Ligands: DGT, MG, GOL |
 |
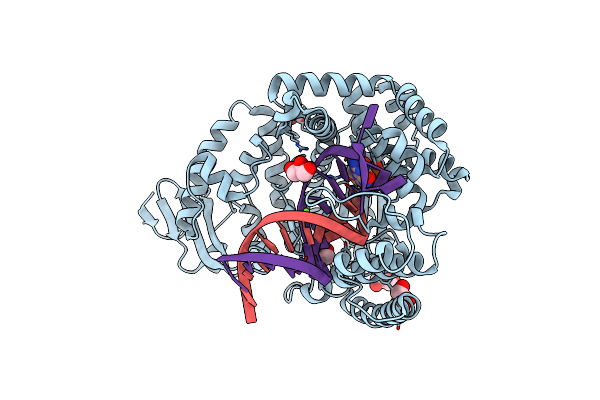
Structure Of Large Fragment Of Dna Polymerase I From Thermus Aquaticus Host-Guest Complex With The Unnatural Base M-Fc Pair With Dg
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2018-11-21 Classification: TRANSFERASE/DNA Ligands: DGT, MG, GOL |
 |
Structure Of Large Fragment Of Dna Polymerase I From Thermus Aquaticus Host-Guest Complex With The Unnatural Base 5Fc Pair With Da
Organism: Thermus aquaticus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2018-11-21 Classification: REPLICATION/DNA Ligands: MG, DGT, GOL |
 |
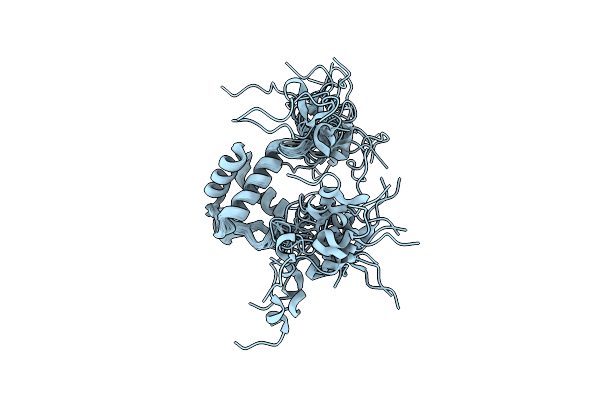
Organism: Bacillus subtilis, Bacillus subtilis subsp. subtilis str. 168
Method: SOLUTION NMR Release Date: 2018-10-17 Classification: DNA BINDING PROTEIN/DNA |
 |
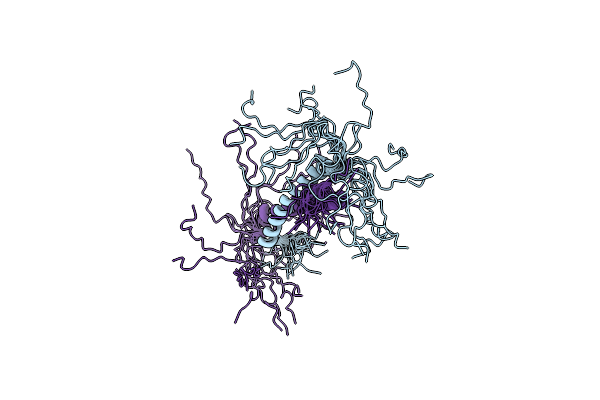
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: SOLUTION NMR Release Date: 2018-10-17 Classification: DNA BINDING PROTEIN |
 |
 |
1.8 Angstrom Crystal Structure Of Succinate-Acetate Permease From Citrobacter Koseri
Organism: Citrobacter koseri (strain atcc baa-895 / cdc 4225-83 / sgsc4696)
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2018-04-25 Classification: TRANSPORT PROTEIN Ligands: ACT, PLM, 78M |

