Search Count: 1,223
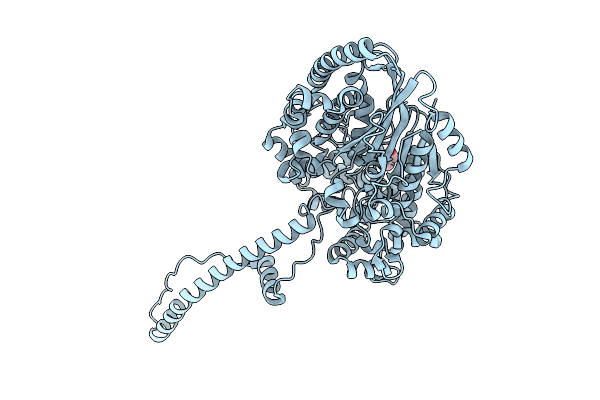 |
Organism: Xanthomonas oryzae pv. oryzae pxo99a
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-08-06 Classification: TOXIN Ligands: GOL |
 |
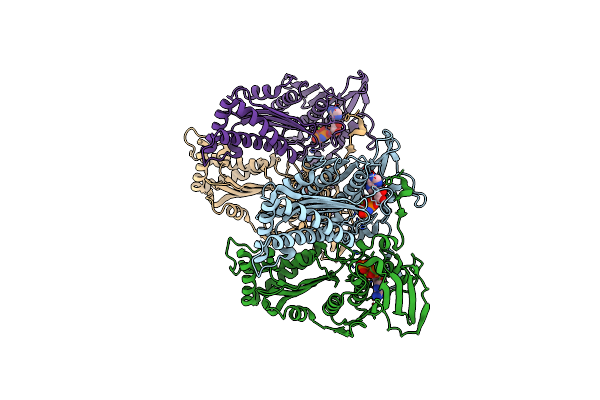
Crystal Structure Of Xanthomonas Oryzae Pv. Oryzae Nadh-Quinone Oxidoreductase Subunits Nuoe And Nuof
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: GOL, FMN, FES |
 |
Organism: Xanthomonas oryzae pv. oryzae (strain kacc10331 / kxo85)
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2021-08-04 Classification: ANTIBIOTIC Ligands: MG, ANP |
 |
Crystal Structure Of Cystathionine Gamma Synthase From Xanthomonas Oryzae Pv. Oryzae In Complex With Homolanthionine
Organism: Xanthomonas oryzae pv. oryzae kacc 10331
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2020-12-09 Classification: BIOSYNTHETIC PROTEIN Ligands: EBO |
 |
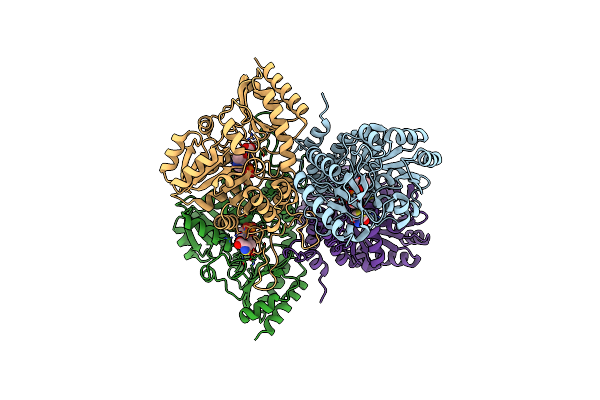
Native Structure Of Cystathionine Gamma Synthase (Xometb) From Xanthomonas Oryzae Pv. Oryzae
Organism: Xanthomonas oryzae pv. oryzae kacc 10331
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, PGE |
 |
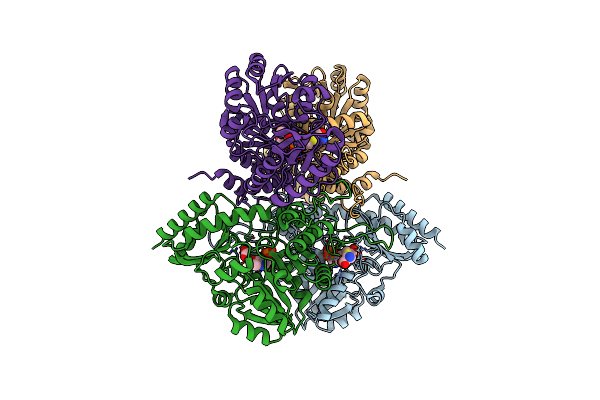
Crystal Structure Of Cystathionine Gamma Synthase From Xanthomonas Oryzae Pv. Oryzae In Complex With Aminoacrylate And Cysteine
Organism: Xanthomonas oryzae pv. oryzae (strain kacc10331 / kxo85)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN Ligands: 0JO, CYS |
 |
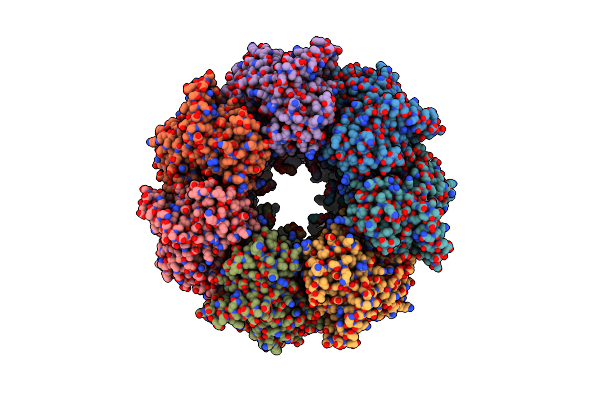
Crystal Structure Of Cystathionine Gamma Synthase From Xanthomonas Oryzae Pv. Oryzae In Complex With Cystathionine
Organism: Xanthomonas oryzae pv. oryzae kacc 10331
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-11-25 Classification: BIOSYNTHETIC PROTEIN Ligands: E9U |
 |
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:3.22 Å Release Date: 2020-07-08 Classification: CHAPERONE Ligands: GOL, SO4 |
 |
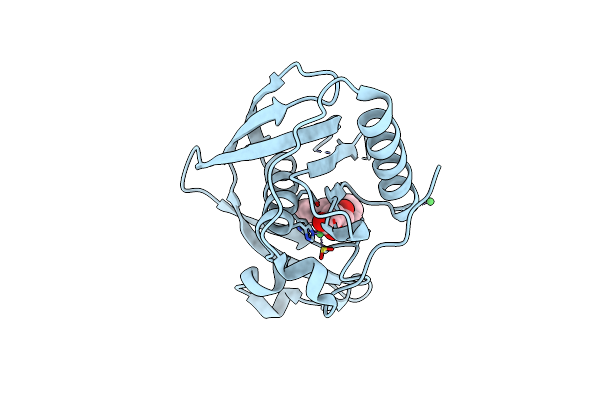
K1U Complex Structure Of Peptide Deformylase From Xanthomonas Oryzae Pv. Oryzae
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-10-23 Classification: HYDROLASE Ligands: CD, NI, K1U |
 |
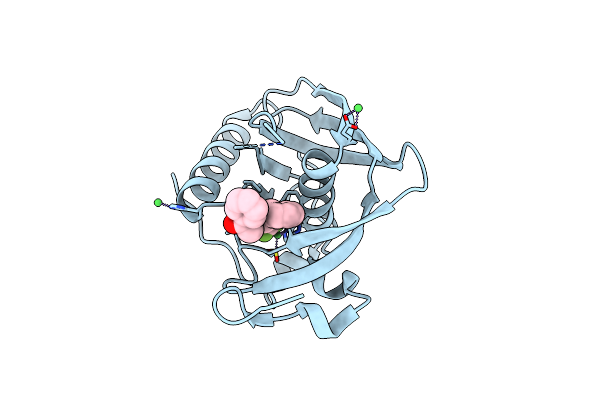
K2U Complex Structure Of Peptide Deformylase From Xanthomonas Oryzae Pv. Oryzae
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-10-23 Classification: HYDROLASE Ligands: NI, K2U |
 |
K3U Complex Structure Of Peptide Deformylase From Xanthomonas Oryzae Pv. Oryzae
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2019-10-23 Classification: HYDROLASE Ligands: K3U, NI |
 |
K4U Complex Structure Of Peptide Deformylase From Xanthomonas Oryzae Pv. Oryzae
Organism: Xanthomonas oryzae pv. oryzae (strain kacc10331 / kxo85)
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2019-10-23 Classification: HYDROLASE Ligands: CD, LHY |
 |
Cryo-Em Structure Of Xanthomonos Oryzae Transcription Elongation Complex With Nusa And The Bacteriophage Protein P7
Organism: Xanthomonas oryzae pv. oryzae (strain pxo99a), Xanthomonas oryzae pv. oryzae pxo99a, Xanthomonas oryzae pv. oryzae, Xanthomonas virus xp10, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2019-07-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo-Em Structure Of Xanthomonos Oryzae Transcription Elongation Complex With The Bacteriophage Protein P7
Organism: Xanthomonas oryzae pv. oryzae pxo99a, Xanthomonas oryzae pv. oryzae maff 311018, Xanthomonas oryzae pv. oryzae, Xanthomonas virus xp10, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2019-07-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Crystal Structure Of Catalytic Domain Of 1,4-Beta-Cellobiosidase (Cbsa) From Xanthomonas Oryzae Pv. Oryzae
Organism: Xanthomonas oryzae pv. oryzae
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2018-05-16 Classification: HYDROLASE |
 |
Structure Of Glutamyl-Trna Synthetase (Xoo1504) From Xanthomonas Oryzae Pv. Oryzae
Organism: Xanthomonas oryzae pv. oryzae (strain kacc10331 / kxo85)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-10-11 Classification: LIGASE |
 |
Native Structure Of Xoo1075, A Peptide Deformylase From Xanthomonas Oryzae Pv. Oryzae
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-10-12 Classification: HYDROLASE Ligands: CD |
 |
Mas Complex Structure Of Peptide Deformylase From Xanthomonas Oryzae Pv Oryzae
Organism: Xanthomonas oryzae pv. oryzae kacc10331, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-08-03 Classification: HYDROLASE/SUBSTRATE Ligands: CD, ACT, NA |
 |
Methionine-Alanine Complex Structure Of Peptide Deformylase From Xanthomonas Oryzae Pv. Oryzae
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-08-03 Classification: HYDROLASE Ligands: CD, ACT, NA, GOL, MET, ALA |
 |
Structure Of Xoo1075, A Peptide Deformyase From Xanthomonas Oryzae Pv. Oryze, In Complex With Fragment 493
Organism: Xanthomonas oryzae pv. oryzae kacc10331
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-08-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CD, 56E, ACT, NA |

