Search Count: 291
All
Selected
 |
Crystal Structure Of Xanthobacter Autotrophicus Sparda Mutant Lacking Dren Nuclease Domains
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN Ligands: ACY, GOL, TRS, PEG, CL, BME, TAR |
 |
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN Ligands: SO4 |
 |
Organism: Xanthobacter autotrophicus py2, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of F501H/H506E Variant Of 2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase (2-Kpcc) From Xanthobacter Autotrophicus
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: COM, FDA, MG |
 |
Crystal Structure Of F501H Variant Of 2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase (2-Kpcc) From Xanthobacter Autotrophicus
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-07-21 Classification: OXIDOREDUCTASE Ligands: FDA, COM, MG |
 |
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2017-08-09 Classification: LIGASE Ligands: MN, ACT, AE4, MG, AMP, ZN |
 |
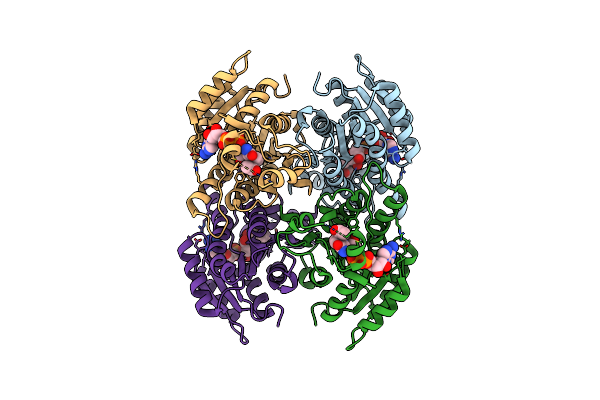
Mechanism Of Atp-Dependent Acetone Carboxylation, Acetone Carboxylase Amp Bound Structure
Organism: Xanthobacter autotrophicus (strain atcc baa-1158 / py2)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-08-09 Classification: LIGASE Ligands: MN, AMP, SO4, ZN |
 |
Mechanism Of Atp-Dependent Acetone Carboxylation, Acetone Carboxylase Nucleotide-Free Structure
Organism: Xanthobacter autotrophicus (strain atcc baa-1158 / py2)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-08-09 Classification: LIGASE Ligands: MN, SO4, ZN |
 |
Crystal Structure Of D-Isomer Specific 2-Hydroxyacid Dehydrogenase From Xanthobacter Autotrophicus Py2 In Complex With Nadph And Mes.
Organism: Xanthobacter autotrophicus (strain atcc baa-1158 / py2)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-04-26 Classification: OXIDOREDUCTASE Ligands: NDP, MES |
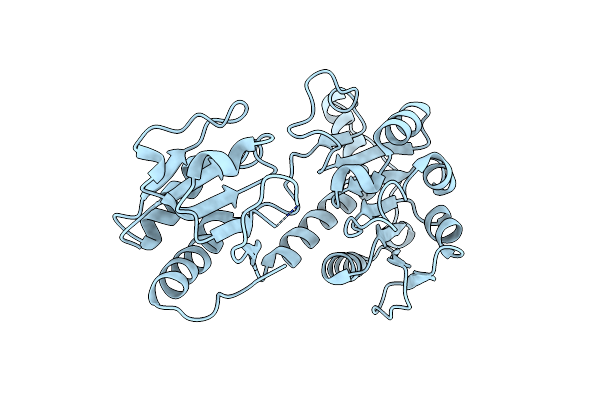 |
Crystal Structure Of D-Isomer Specific 2-Hydroxyacid Dehydrogenase From Xanthobacter Autotrophicus Py2
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-01-14 Classification: OXIDOREDUCTASE |
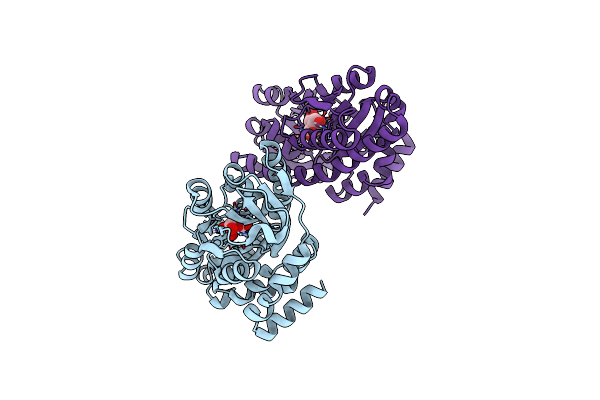 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Xanthobacter Autotrophicus Py2, Target Efi-510329, With Bound Beta-D-Galacturonate
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-01-22 Classification: SOLUTE-BINDING PROTEIN Ligands: CL, GTR |
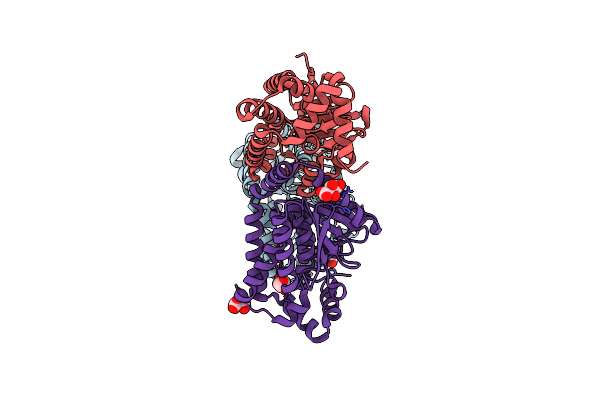 |
Crystal Structure Of An Enoyl-Coa Hydratase/Isomerase From Xanthobacter Autotrophicus Py2
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2013-04-24 Classification: ISOMERASE Ligands: TLA, GOL |
 |
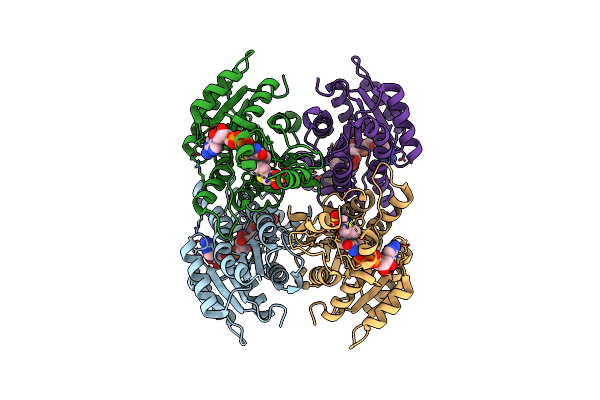
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-03-27 Classification: OXIDOREDUCTASE Ligands: NAD, ACT |
 |
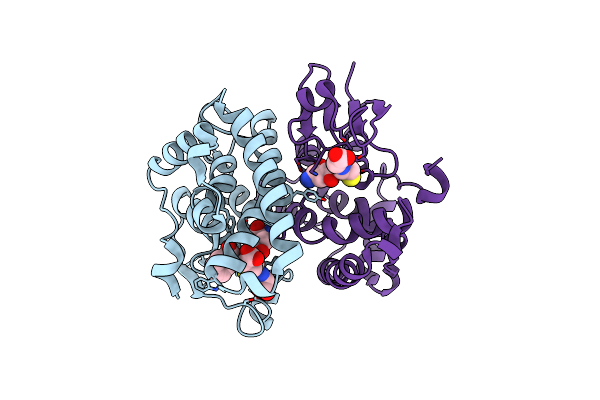
Crystal Structure Of S-2-Hydroxypropyl Coenzyme M Dehydrogenase (S-Hpcdh) Bound To S-Hpc And Nadh
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-03-27 Classification: OXIDOREDUCTASE Ligands: NAI, 1HS |
 |
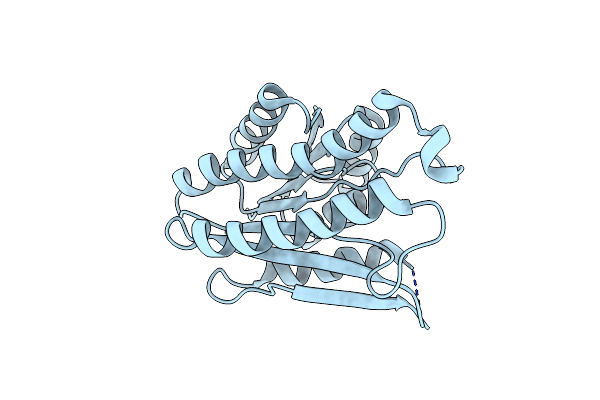
Crystal Structure Of Glutathione S-Transferase Xaut_3756 (Target Efi-507152) From Xanthobacter Autotrophicus Py2
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-11-28 Classification: TRANSFERASE Ligands: GSH, BEZ, UNL |
 |
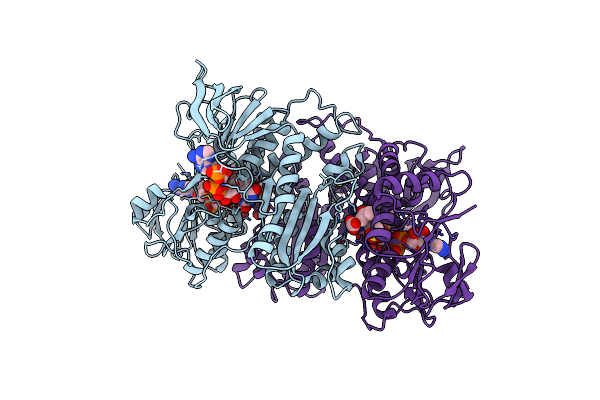
Crystal Structure Of A Short-Chain Dehydrogenase/Reductase Sdr From Xanthobacter Autotrophicus Py2
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-03-14 Classification: OXIDOREDUCTASE Ligands: CL |
 |
Structural Basis For Carbon Dioxide Binding By 2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2011-02-16 Classification: OXIDOREDUCTASE Ligands: NAP, FAD, KPC, MG, BCT, CO2, ACN, COM |
 |
The Effect Of Deuteration On Protein Structure A High Resolution Comparison Of Hydrogenous And Perdeuterated Haloalkane Dehalogenase
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2007-09-04 Classification: HYDROLASE |
 |
The Effect Of Deuteration On Protein Structure A High Resolution Comparison Of Hydrogenous And Perdeuterated Haloalkane Dehalogenase
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2007-09-04 Classification: HYDROLASE |
 |
Structural Basis For Stereo Selectivity In The (R)- And (S)- Hydroxypropylethane Thiosulfonate Dehydrogenases
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-07-26 Classification: OXIDOREDUCTASE Ligands: NAD, KPC |

