Search Count: 10
 |
Organism: Actinomycetota bacterium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-02-28 Classification: FLAVOPROTEIN Ligands: FAD, XUL |
 |
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With Two Co2+ Ions And Xylulose
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: CO, XUL, HYP |
 |
Crystal Structure Of Human D-Xylulokinase In Complex With D-Xylulose And Adenosine Diphosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2012-11-28 Classification: TRANSFERASE Ligands: ADP, XUL, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2012-11-28 Classification: TRANSFERASE Ligands: XUL, EDO |
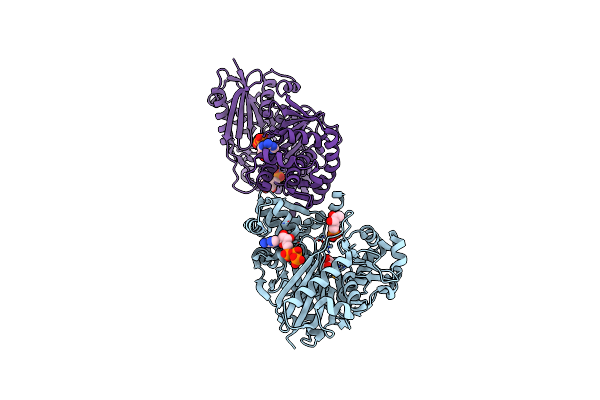 |
The Crystal Structure Of Ligand Bound Xylulose Kinase From Lactobacillus Acidophilus
Organism: Lactobacillus acidophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-03-23 Classification: TRANSFERASE Ligands: ATP, DXP, XUL, ADP |
 |
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2010-01-05 Classification: TRANSFERASE Ligands: XUL, EPE, GOL |
 |
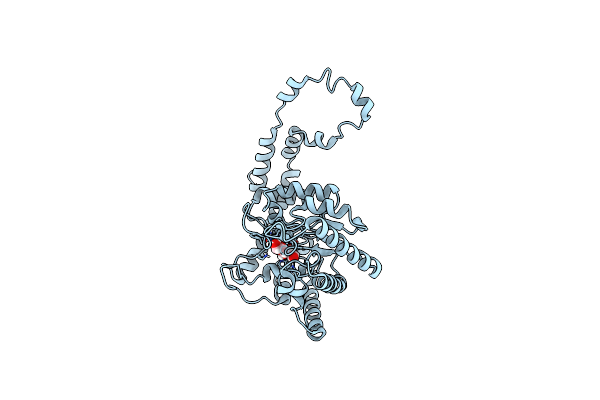
Organism: Chromobacterium violaceum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2009-07-21 Classification: TRANSFERASE Ligands: ADP, PO4, GOL, MLI, XUL |
 |
Organism: Streptomyces rubiginosus
Method: NEUTRON DIFFRACTION Resolution:2.20 Å Release Date: 2008-08-05 Classification: ISOMERASE Ligands: XUL, MG, OH, DOD |
 |
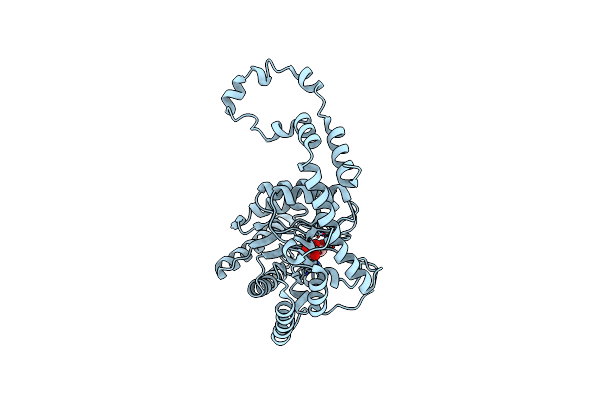
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-11-14 Classification: TRANSFERASE Ligands: XUL, SO4, NH4 |
 |
Modes Of Binding Substrates And Their Analogues To The Enzyme D-Xylose Isomerase
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1994-06-22 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: XUL, MN |

