Search Count: 21
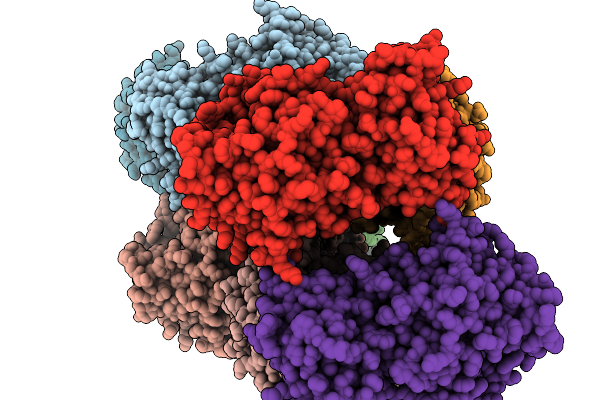 |
Crystal Structure Of Pseudopedobacter Saltans Gh43 Beta-Xylosidase In Complex With Xylose.
Organism: Pseudopedobacter saltans dsm 12145
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2026-01-07 Classification: HYDROLASE Ligands: CA, XLS |
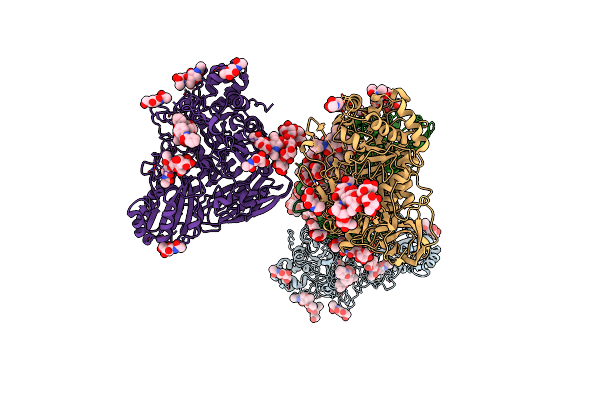 |
Bifunctional Xylosidase/Glucosidase Lxyl With Intermediate Substrate Xylose, 120 Seconds
Organism: Lentinula edodes
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-10-25 Classification: HYDROLASE Ligands: BKR, XLS, MAN, NAG, BMA |
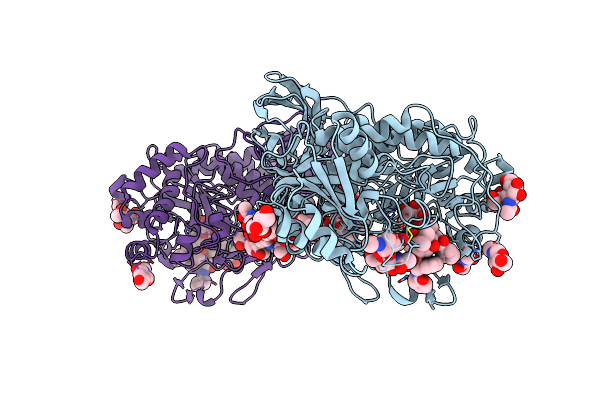 |
Bifunctional Xylosidase/Glucosidase Lxyl With Intermediate Substrate Xylose, 5 Seconds
Organism: Lentinula edodes
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2023-08-09 Classification: HYDROLASE Ligands: NAG, BKR, XLS |
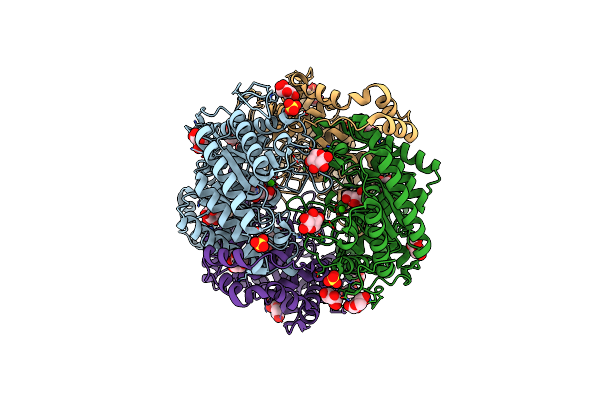 |
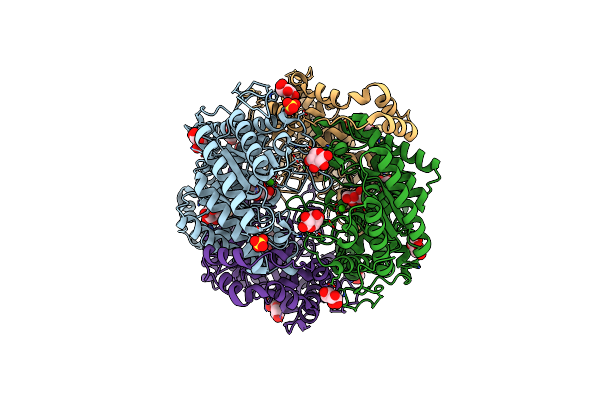
Crystal Structure Of Native Xylose Isomerase From Piromyces E2 Grown In Yeast, In Complex With Xylose
Organism: Piromyces sp. (strain e2)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2020-01-29 Classification: ISOMERASE Ligands: CA, XLS, XYP, XYS, SO4 |
 |
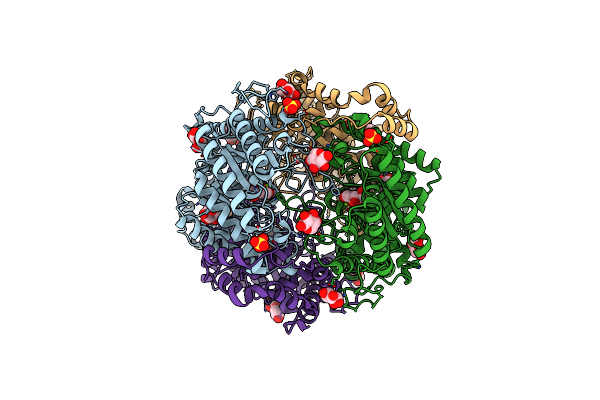
Crystal Structure Of Mutant Xylose Isomerase (V270A/A273G) From Piromyces E2 Grown In Yeast, In Complex With Xylose
Organism: Piromyces sp. (strain e2)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-01-29 Classification: ISOMERASE Ligands: CA, XLS, XYP, XYS, SO4 |
 |
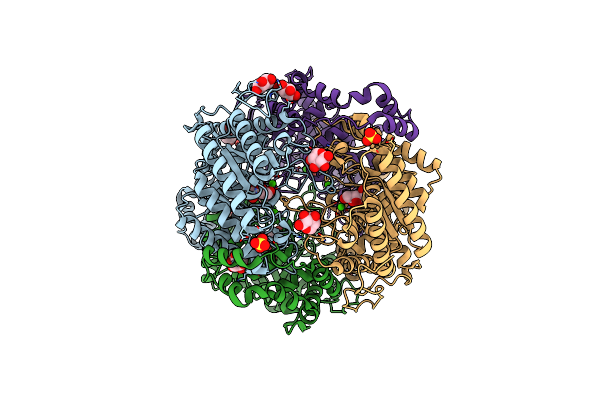
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With Two Mg2+ Ions And Xylose
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: MG, XLS, XYP, XYS, SO4 |
 |
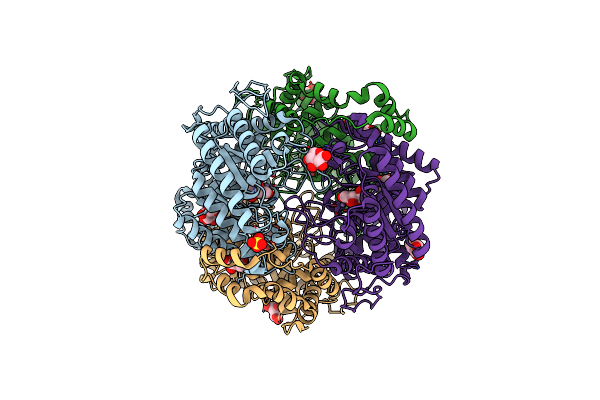
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With Two Ca2+ Ions And Xylose
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: CA, XLS, XYP, SO4 |
 |
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With Two Mn2+ Ions And Xylose
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: MN, XLS, XYP, SO4 |
 |
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With 2 Ni2+ Ions And Xylose
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: NI, XLS, XYP, XYS, SO4 |
 |
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With Two Cd2+ Ions And Xylose
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: CD, XLS, XYP, XYS, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2011-01-12 Classification: TOXIN Ligands: XLS |
 |
Crystal Structure Of A Putative Sugar Phosphate Isomerase/Epimerase (Ava4194) From Anabaena Variabilis Atcc 29413 At 1.78 A Resolution
Organism: Anabaena variabilis atcc 29413
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2007-09-04 Classification: ISOMERASE Ligands: XLS, ZN, CL, GOL |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-04-24 Classification: LYASE Ligands: SIE, XLS, SO4 |
 |
Modes Of Binding Substrates And Their Analogues To The Enzyme D-Xylose Isomerase
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1994-06-22 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: XLS, MN |
 |
Protein Engineering Of Xylose (Glucose) Isomerase From Actinoplanes Missouriensis. 1. Crystallography And Site-Directed Mutagenesis Of Metal Binding Sites
Organism: Actinoplanes missouriensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1993-07-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: XLS, MG |
 |
Protein Engineering Of Xylose (Glucose) Isomerase From Actinoplanes Missouriensis. 1. Crystallography And Site-Directed Mutagenesis Of Metal Binding Sites
Organism: Actinoplanes missouriensis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1993-07-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: XLS, MG |
 |
Protein Engineering Of Xylose (Glucose) Isomerase From Actinoplanes Missouriensis. 1. Crystallography And Site-Directed Mutagenesis Of Metal Binding Sites
Organism: Actinoplanes missouriensis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1993-07-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: XLS, MG |
 |
Protein Engineering Of Xylose (Glucose) Isomerase From Actinoplanes Missouriensis. 1. Crystallography And Site-Directed Mutagenesis Of Metal Binding Sites
Organism: Actinoplanes missouriensis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1993-07-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: XLS, MN |
 |
A Metal-Mediated Hydride Shift Mechanism For Xylose Isomerase Based On The 1.6 Angstroms Streptomyces Rubiginosus Structures With Xylitol And D-Xylose
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1992-07-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: XYS, XLS, MG |
 |
A Metal-Mediated Hydride Shift Mechanism For Xylose Isomerase Based On The 1.6 Angstroms Streptomyces Rubiginosus Structures With Xylitol And D-Xylose
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 1992-07-15 Classification: ISOMERASE(INTRAMOLECULAR OXIDOREDUCTASE) Ligands: XLS, XYS, MN |

