Search Count: 145
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: LDA, ACY, GOL, XE, SO4, NH2 |
 |
Bovine Heart Cytochrome C Oxidase In The Xenon-Bound Fully Oxidized State Under Aerobic Condition
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, CDL, LFA, DMU, EDO, XE, PGV, CUA, CHD, UNX, ZN, PEK |
 |
Bovine Heart Cytochrome C Oxidase In The Xenon-Bound Fully Oxidized State Under Anaerobic Condition
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, CDL, LFA, DMU, EDO, XE, PGV, CUA, CHD, UNX, ZN, PEK |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, CDL, LFA, DMU, EDO, XE, PGV, CUA, CHD, UNX, ZN, PEK |
 |
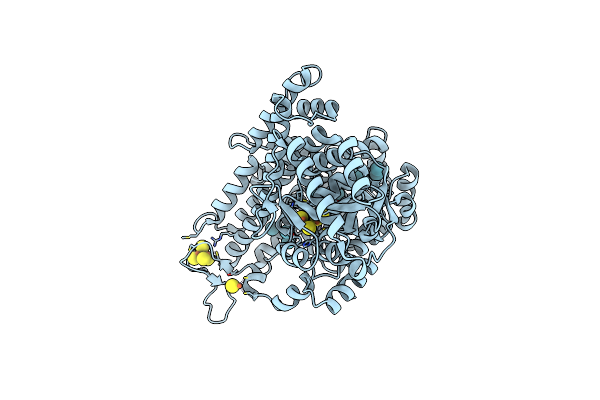
Room Temperature Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) Under Xenon Pressure (30 Bar)
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-07-05 Classification: OXIDOREDUCTASE Ligands: XE, TAR |
 |
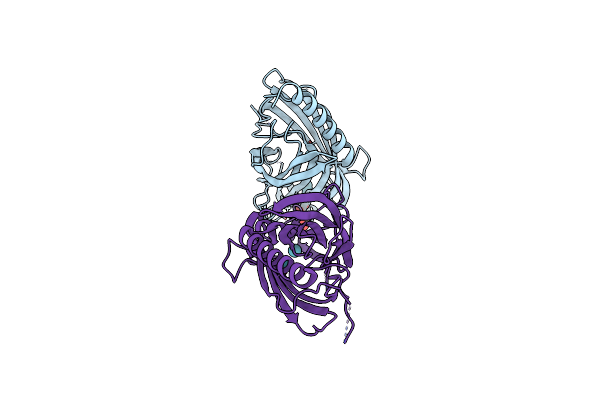
Xenon-Bound Structure Of Carbon Monoxide Dehydrogenase (Codh) From Desulfovibrio Vulgaris
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-05-04 Classification: OXIDOREDUCTASE Ligands: SF4, FES, CUV, XE |
 |
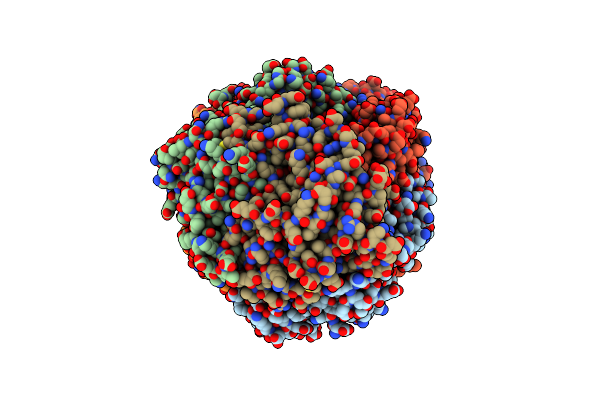
Organism: Mycolicibacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2022-04-13 Classification: UNKNOWN FUNCTION Ligands: XE, GOL |
 |
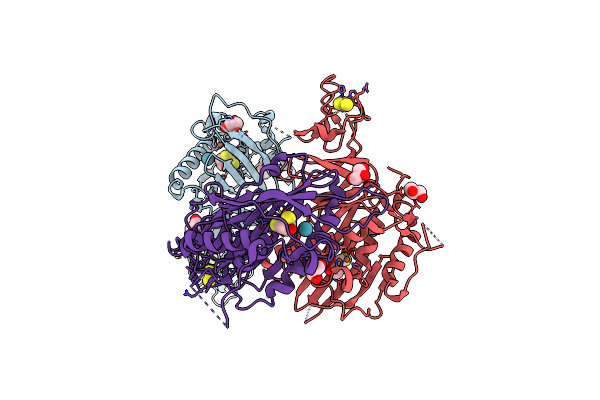
Structure Of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
Organism: Methanothermobacter marburgensis str. marburg
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-04-06 Classification: TRANSFERASE Ligands: F43, MG, ACT, TP7, COM, NA, XE |
 |
Structure Of The Rieske Non-Heme Iron Oxygenase Gxta Pressurized With Xenon
Organism: Microseira wollei
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2022-01-19 Classification: BIOSYNTHETIC PROTEIN Ligands: FES, GOL, FE, XE, DTT, CL |
 |
Organism: Carboxydothermus hydrogenoformans (strain atcc baa-161 / dsm 6008 / z-2901)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: SF4, XE, BR, T2N, H2S, FES |
 |
Crystal Structure Of The Ethyl-Coenzyme M Reductase From Candidatus Ethanoperedens Thermophilum Gassed With Xenon
Organism: Candidatus ethanoperedens thermophilum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-07-14 Classification: TRANSFERASE Ligands: USN, GOL, NA, TP7, COM, K, XE, CL, UWT, MG, TRS |
 |
Crystal Structure Of The Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis Derivatized With Xenon
Organism: Methanothermobacter marburgensis (strain atcc baa-927 / dsm 2133 / jcm 14651 / nbrc 100331 / ocm 82 / marburg)
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2021-07-14 Classification: TRANSFERASE Ligands: COM, TP7, K, PEG, GOL, EDO, F43, MG, XE, SO4, CL |
 |
Organism: Xenopus tropicalis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-11-11 Classification: UNKNOWN FUNCTION Ligands: XE |
 |
Xenon Derivatization Of The F420-Reducing [Nife] Hydrogenase Complex From Methanosarcina Barkeri
Organism: Methanosarcina barkeri ms
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2019-10-23 Classification: OXIDOREDUCTASE Ligands: SF4, 144, FES, XE, BU3, NFU, FE, MG, FAD |
 |
Crystal Structure Of Tem1 Beta-Lactamase Mutant I263A In The Presence Of 1.2 Mpa Xenon
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2018-09-12 Classification: HYDROLASE Ligands: XE |
 |
E. Coli Phosphoenolpyruvate Carboxykinase G209S K212C Mutant Bound To Thiosulfate
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-08-29 Classification: LYASE Ligands: ATP, MG, MN, XE, BTB, THJ |
 |
Organism: Streptomyces maritimus
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2018-05-02 Classification: FLAVOPROTEIN Ligands: FAD, XE |
 |
Crystal Structure Of Murine Neuroglobin Mutant V140W Under 20 Bar Xenon Pressure
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-11-15 Classification: TRANSPORT PROTEIN Ligands: HEM, SO4, XE, DIO |
 |
Crystal Structure Of Murine Neuroglobin Mutant V140W Under 30 Bar Xenon Pressure
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2017-11-15 Classification: TRANSPORT PROTEIN Ligands: HEM, SO4, XE |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-09-20 Classification: MEMBRANE PROTEIN Ligands: PLM, XE, BOG, NA, NI |

