Search Count: 32
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: SF4, FES, XCC, FE |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: SF4, FES, XCC, FE |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: SF4, FES, XCC, FE |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: SF4, FES, XCC, FE |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: SF4, FES, XCC, FE |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: SF4, FES, XCC, FE |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: SF4, FES, XCC, FE |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: SF4, FES, XCC, FE |
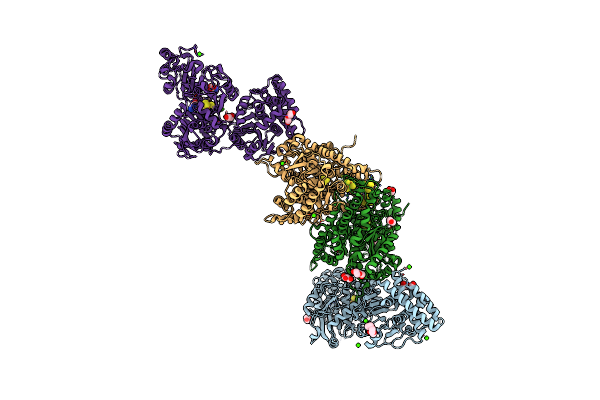 |
Structure Of Carbon Monoxide Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) In Complex With Acetyl-Coenyzme A From Clostridium Autoethanogenum
Organism: Clostridium autoethanogenum dsm 10061
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: SF4, NI, EDO, PE4, GOL, PEG, CA, CL, XCC, ACO, TRS |
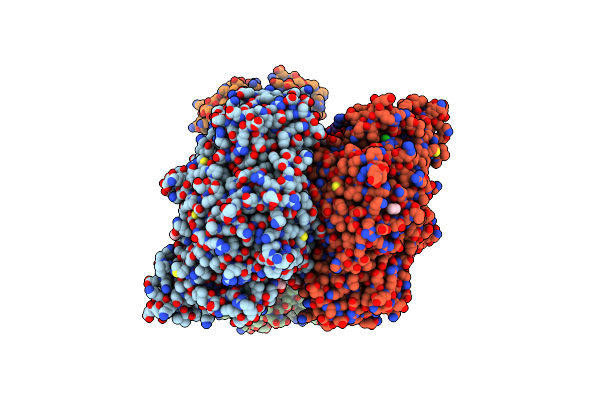 |
Crystal Structure Of The F420-Reducing Carbon Monoxide Dehydrogenase Component From The Ethanotroph Candidatus Ethanoperedens Thermophilum
Organism: Candidatus methanoperedenaceae archaeon gb50
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-10-02 Classification: OXIDOREDUCTASE Ligands: GOL, EDO, SF4, XCC, K, FAD, PE4, CL |
 |
Crystal Structure Of Co Dehydrogenase Mutant With Increased Affinity For Electron Mediators In High Peg Concentration
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE, 1PE |
 |
Crystal Structure Of Co Dehydrogenase Mutant With Increased Affinity For Electron Mediators In Low Peg Concentration
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE, EDO |
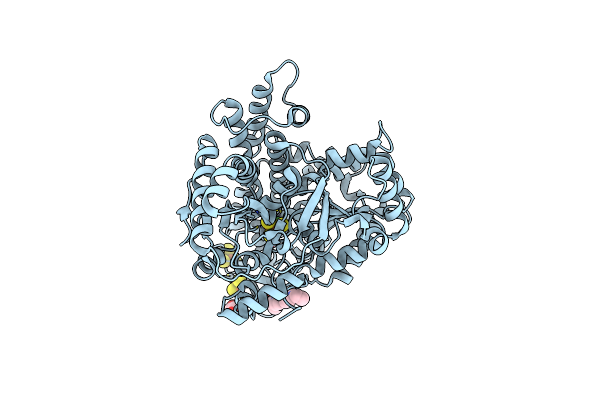 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, S8I, EDO |
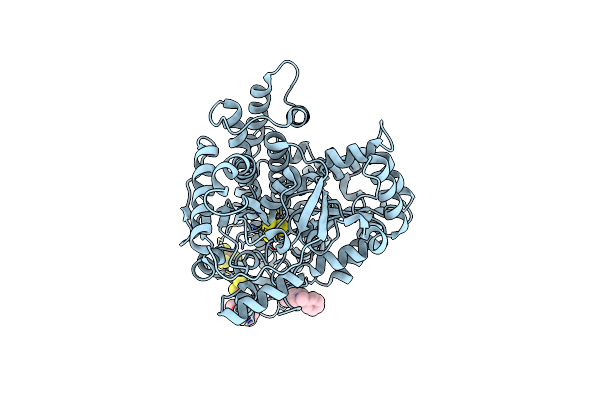 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, PGE, S7L |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-04-17 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, XCC, FE |
 |
Co-Dehydrogenase/Acetyl-Coa Synthase (Codh/Acs) From Clostridium Autoethanogenum At 3.0-A Resolution
Organism: Clostridium autoethanogenum dsm 10061
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2020-11-04 Classification: OXIDOREDUCTASE Ligands: GOL, PE4, SF4, F, NI, XCC, NA |
 |
Co-Dehydrogenase Homodimer From Clostridium Autoethanogenum At 1.90-A Resolution
Organism: Clostridium autoethanogenum dsm 10061
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-11-04 Classification: OXIDOREDUCTASE Ligands: ACT, SF4, XCC, EDO, CL |
 |
Co-Dehydrogenase Coupled To The N-Terminal Domain Of The Acetyl-Coa Synthase From Clostridium Autoethanogenum Isolated After Tryptic Digestion.
Organism: Clostridium autoethanogenum dsm 10061
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2020-11-04 Classification: OXIDOREDUCTASE Ligands: MG, SF4, XCC, GOL, EDO |
 |
Organism: Moorella thermoacetica
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2020-10-07 Classification: OXIDOREDUCTASE/TRANSFERASE Ligands: SF4, XCC, PGE, PG5, NI, CMO, MG |
 |
Crystal Structure Of C45G/T50C D. Vulgaris Carbon Monoxide Dehydrogenase (Anaerobic)
Organism: Desulfovibrio vulgaris (strain hildenborough / atcc 29579 / dsm 644 / ncimb 8303)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2020-07-29 Classification: OXIDOREDUCTASE Ligands: SF4, XCC, CUV, GOL |

