Search Count: 73
 |
Organism: Saccharomyces cerevisiae, Inovirus m13
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA |
 |
Cryo-Em Structure Of Yeast Mgm101 Bound To Duplex Dna Annealing Intermediate
Organism: Saccharomyces cerevisiae, Inovirus m13
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Saccharomyces cerevisiae, Inovirus m13
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ANTIBIOTIC |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ANTIBIOTIC Ligands: MG |
 |
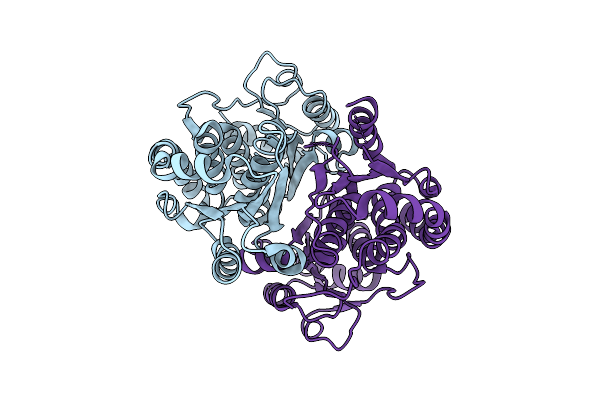
Crystal Structure Of Salmonella Frab Deglycase, Crystal Form 4 With Deletion Of C-Terminal Residues 313-325.
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ANTIBIOTIC |
 |
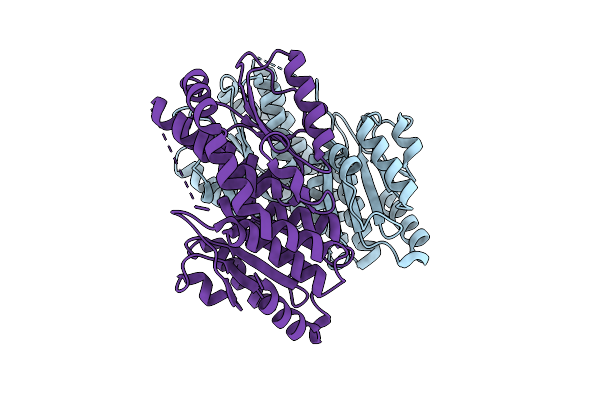
Crystal Structure Of Salmonella Frab Deglycase, E214A Mutant, Crystal Form 5
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ANTIBIOTIC |
 |
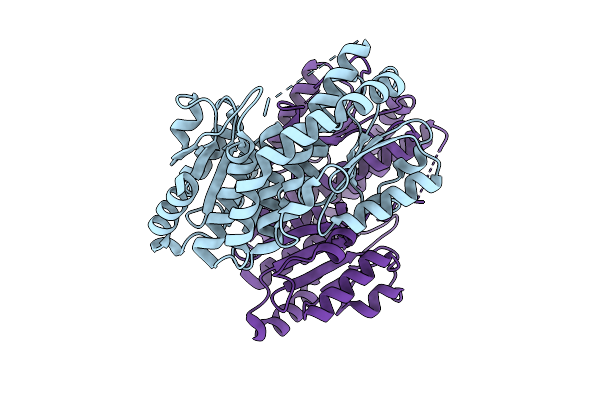
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ANTIBIOTIC |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: ANTIBIOTIC |
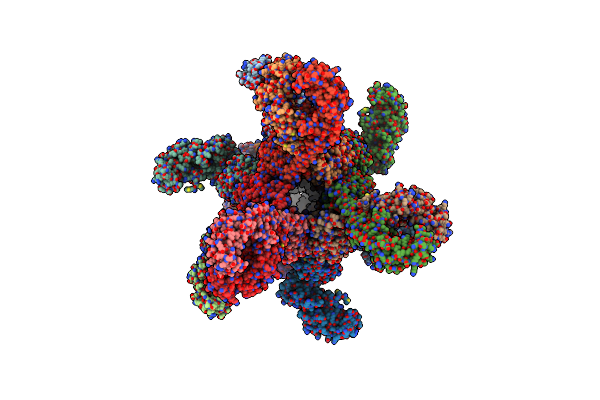 |
Organism: Bacillus sp. hmf5848
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Bacillus sp. hmf5848
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Bacillus sp. hmf5848
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Bacillus sp. hmf5848
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: ANTIVIRAL PROTEIN |
 |
Molecular Basis For Hera-Duf Supramolecular Complex In Anti-Phage Defense - Assembly 3
Organism: Bacillus sp. hmf5848
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: ANTIVIRAL PROTEIN |
 |
Sars-Cov-2 Nucleocapsid Dimerization Domain Bound To Fab-Np1E9 And Fab-Np3B4
Organism: Severe acute respiratory syndrome coronavirus 2, Mus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: VIRAL PROTEIN |
 |
Alkalihalobacillus Halodurans (Aha) Trp Rna Binding Attenuation Protein (Trap) Mutant Dtrap With Trp
Organism: Halalkalibacterium halodurans
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: RNA BINDING PROTEIN Ligands: TRP |
 |
Alkalihalobacillus Halodurans (Aha) Trp Rna Binding Attenuation Protein (Trap) Mutant Dtrap Without Trp
Organism: Halalkalibacterium halodurans
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: RNA BINDING PROTEIN |
 |
Alkalihalobacillus Halodurans (Aha) Trp Rna Binding Attenuation Protein (Trap) Mutant T49A/T52A Dtrap With Trp
Organism: Halalkalibacterium halodurans
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: RNA BINDING PROTEIN Ligands: TRP |

