Search Count: 21
 |
Semet Crystal Structure Of Erwinia Amylovora Amyr Amylovoran Repressor, A Member Of The Ybjn Protein Family
Organism: Erwinia amylovora
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2017-02-15 Classification: TRANSCRIPTION |
 |
Erwinia Amylovora Amyr Amylovoran Repressor, A Member Of The Ybjn Protein Family
Organism: Erwinia amylovora
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-01-18 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of Glucose-1-Phosphate Uridylyltransferase Galu From Erwinia Amylovora.
Organism: Erwinia amylovora
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2016-01-20 Classification: TRANSFERASE |
 |
Organism: Erwinia amylovora
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2015-08-05 Classification: TRANSFERASE Ligands: FRU, GLC |
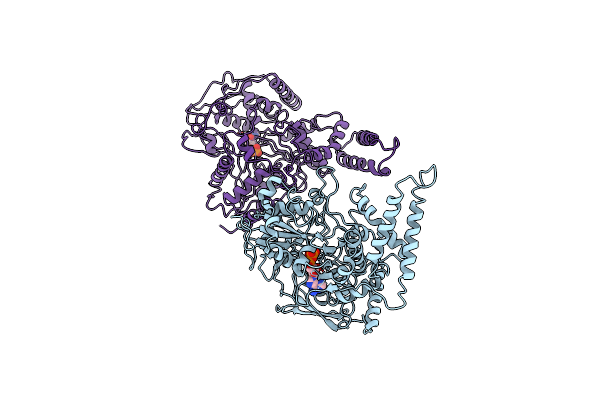
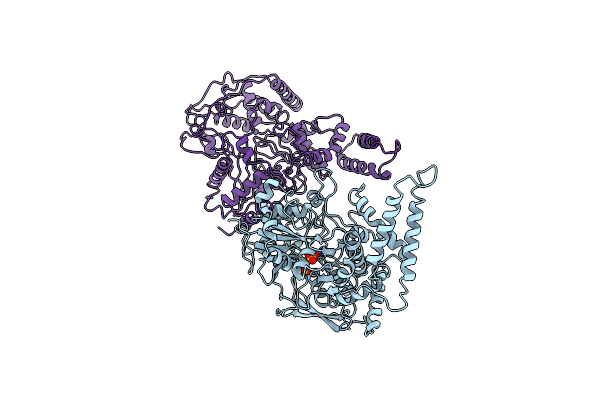 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-12-12 Classification: NUCLEAR PROTEIN Ligands: SO4 |
 |
Adp-Bound C-Terminal Domain Of Actin-Related Protein Arp8 From S. Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2012-12-12 Classification: NUCLEAR PROTEIN Ligands: ADP |
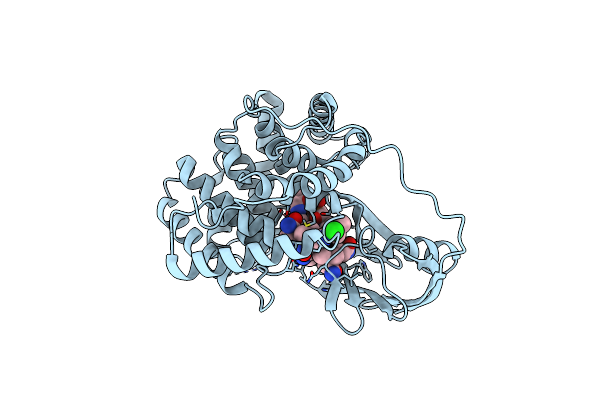 |
Crystallographic Analysis Of Upper Axial Ligand Substitutions In Cobalamin Bound To Transcobalamin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2007-10-30 Classification: TRANSPORT PROTEIN Ligands: B12, CYN, CL |
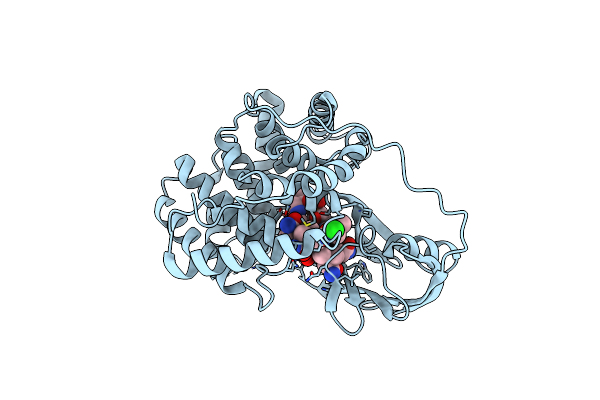 |
Crystallographic Analysis Of Beta-Axial Ligand Substitutions In Cobalamin Bound To Transcobalamin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2007-10-30 Classification: TRANSPORT PROTEIN Ligands: B12, CL, SO3 |
 |
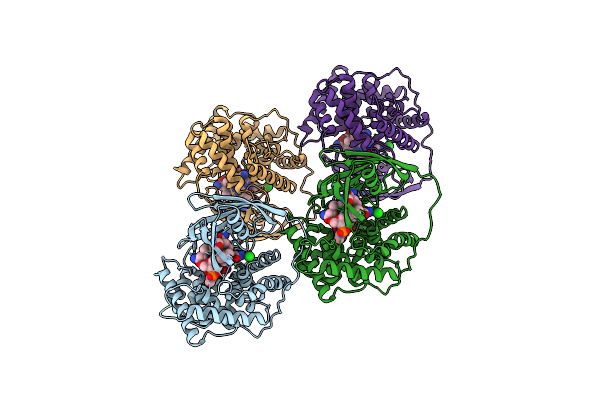
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2006-04-04 Classification: TRANSPORT PROTEIN Ligands: B12 |
 |
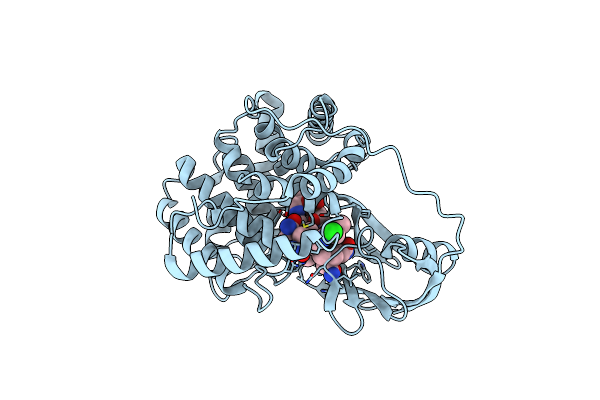
Structure Of Cobalamin-Complexed Bovine Transcobalamin In Monoclinic Crystal Form
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-04-04 Classification: TRANSPORT PROTEIN Ligands: CL, B12 |
 |
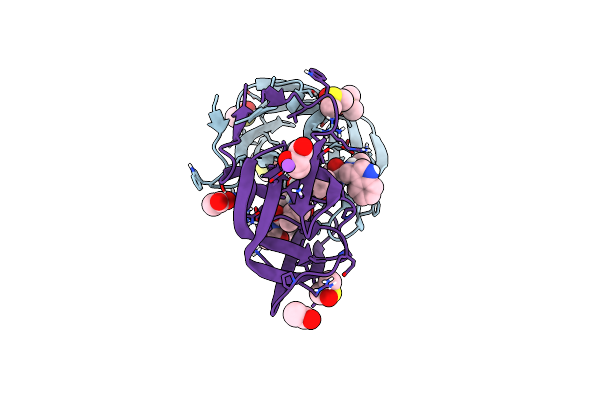
Structure Of Cobalamin-Complexed Bovine Transcobalamin In Trigonal Crystal Form
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-04-04 Classification: TRANSPORT PROTEIN Ligands: CL, B12 |
 |
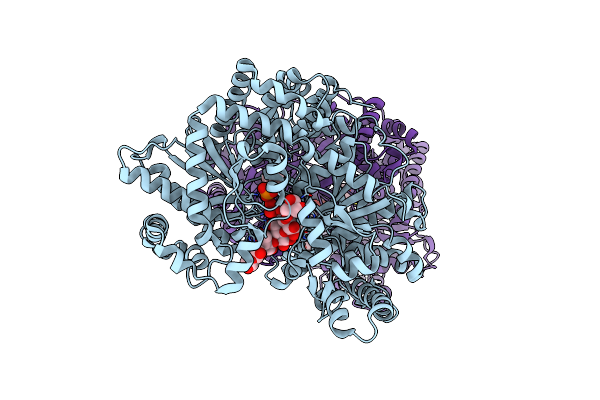
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2006-02-21 Classification: HYDROLASE Ligands: DMS, NA, CL, ACT, IPF, GOL |
 |
X-Ray Studies On Maltodextrin Phosphorylase (Malp) Complexes: Recognition Of Substrates And Catalytic Mechanism Of Phosphorylase Family
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2005-09-20 Classification: TRANSFERASE Ligands: VO4, PLP, TRS |
 |
X-Ray Studies On Maltodextrin Phosphorylase Complexes: Recognition Of Substrates And Cathalitic Mechanism Of Phosphorylase Family
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-09-13 Classification: TRANSFERASE Ligands: SO4, PLP |
 |
X-Ray Studies On Maltodextrin Phosphorylase Complexes: Recognition Of Substrates And Cathalitic Mechanism Of Phosphorylase Family
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2005-09-06 Classification: TRANSFERASE Ligands: NO3, PLP |
 |
Crystal Structure Of Ni-Containing Superoxide Dismutase With Ni-Ligation Corresponding To The Oxidized State
Organism: Streptomyces seoulensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-05-18 Classification: OXIDOREDUCTASE Ligands: SO4, 3NI |
 |
Crystal Structure Of Ni-Containing Superoxide Dismutase With Ni-Ligation Corresponding To The State After Partial X-Ray-Induced Reduction
Organism: Streptomyces seoulensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-05-18 Classification: OXIDOREDUCTASE Ligands: SO4, 3NI |
 |
Crystal Structure Of Ni-Containing Superoxide Dismutase With Ni-Ligation Corresponding To The State After Full X-Ray-Induced Reduction
Organism: Streptomyces seoulensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2004-05-18 Classification: OXIDOREDUCTASE Ligands: NI, SO4 |
 |
Crystal Structure Of Ni-Containing Superoxide Dismutase With Ni-Ligation Corresponding To The Thiosulfate-Reduced State
Organism: Streptomyces seoulensis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-05-18 Classification: OXIDOREDUCTASE Ligands: NI, SO4, THJ |
 |
Crystal Structure Of Ni-Containing Superoxide Dismutase With Ni-Ligation Corresponding To The State After Full X-Ray-Induced Reduction
Organism: Streptomyces seoulensis
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2004-05-18 Classification: OXIDOREDUCTASE Ligands: NI, SO4, ACY |

