Search Count: 366
 |
Organism: Rattus norvegicus, Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: CELL CYCLE,STRUCTURAL PROTEIN/INHIBITOR Ligands: GTP, GDP, A1BMG, SO4, MG |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: LYASE Ligands: FES |
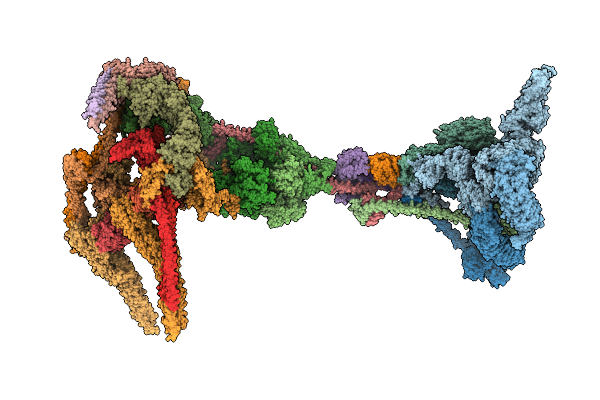 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN Ligands: ZN |
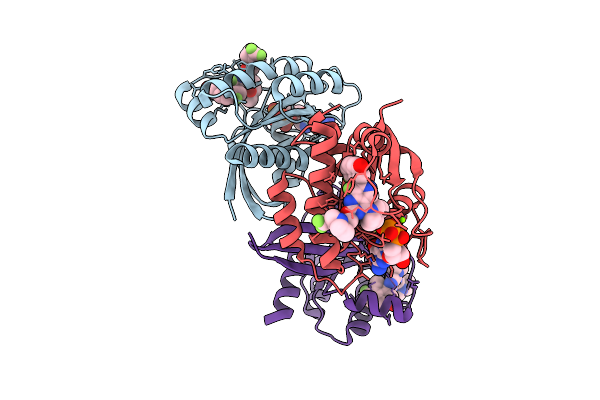 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ONCOPROTEIN Ligands: MG, A1EN3, GDP |
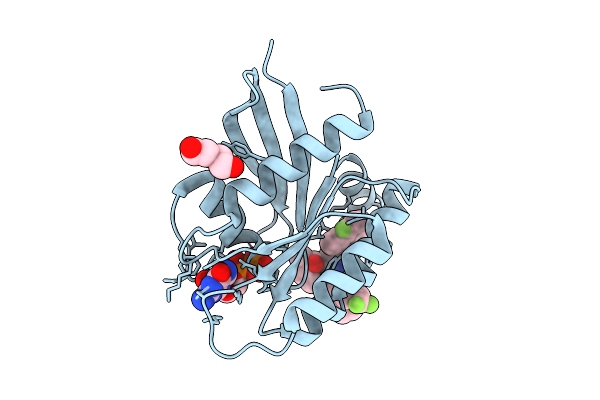 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ONCOPROTEIN Ligands: MG, A1EN3, GDP, PEG |
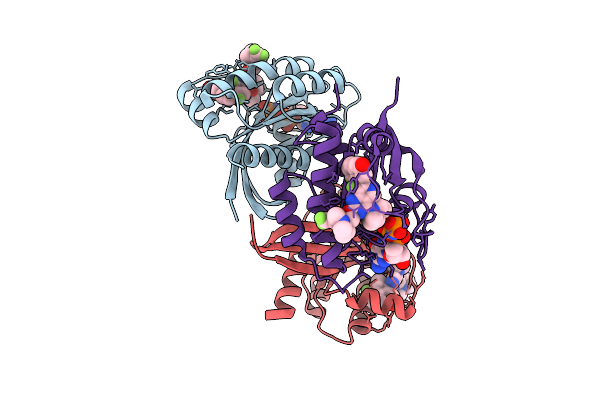 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ONCOPROTEIN Ligands: MG, A1EN3, GNP |
 |
Organism: Kutzneria albida dsm 43870
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: NDP, NA |
 |
Organism: Kutzneria albida dsm 43870
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: NDP, SO4, PE8, NA |
 |
Crystal Structure Of Homolog Of Dihydroxyacid Dehydratase(Astd) From Aspergillus Terreus
Organism: Aspergillus terreus
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: LYASE Ligands: PEG, EDO |
 |
Organism: Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Rothia dentocariosa, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN/DNA/RNA |
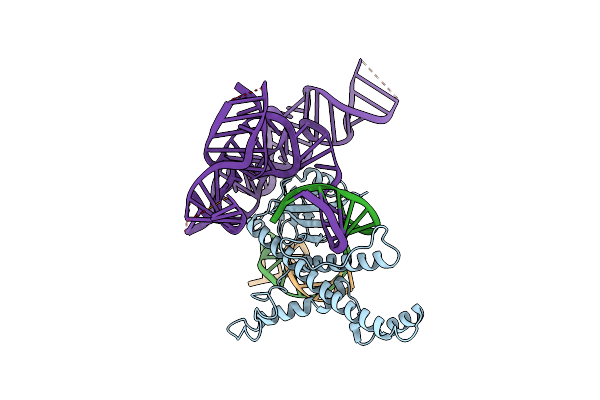 |
Cryo-Em Structure Of The Rdcas12N-Sgrna-Dna Ternary Complex, Conformation 2
Organism: Rothia dentocariosa, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: DNA BINDING PROTEIN/DNA/RNA |
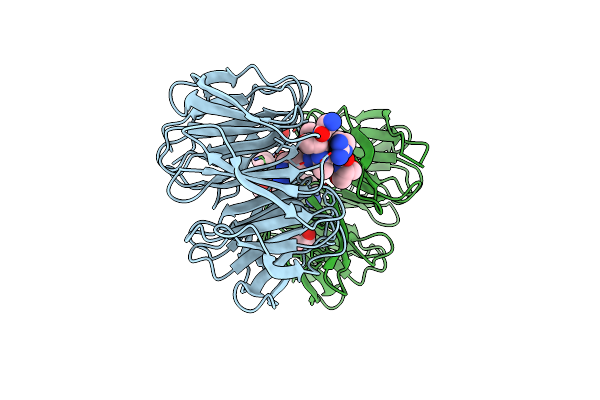 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-05-21 Classification: CYTOSOLIC PROTEIN Ligands: EDO |
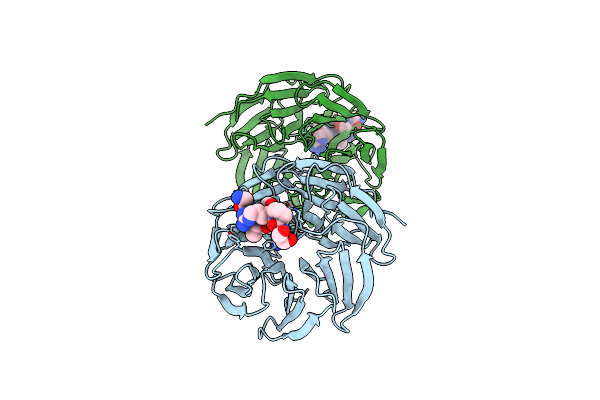 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-21 Classification: NUCLEAR PROTEIN |
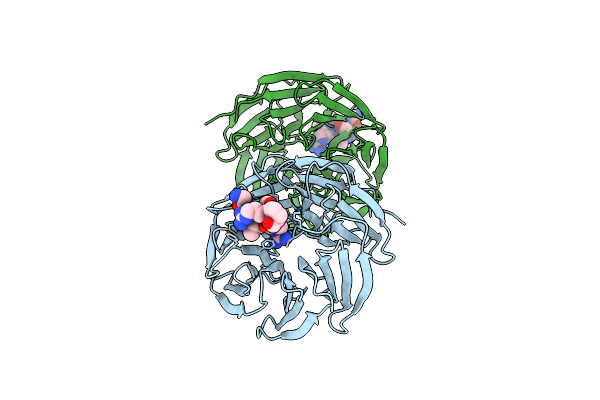 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-05-21 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-21 Classification: NUCLEAR PROTEIN |
 |
Crystal Structure Of The Myb Domain Of S.Pombe Tbf1 In The P222 Space Group
Organism: Schizosaccharomyces pombe 972h-
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-04-09 Classification: DNA BINDING PROTEIN Ligands: GOL |
 |
Cryo-Em Structure Of Nanodisc-Reconstituted Wildtype Human Mrp4 (In Complex With Vincristine)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: R1Q, ANP, MG |
 |
Cryo-Em Structure Of Nanodisc-Reconstituted Human Mrp4 Withe1202Q Mutation (In Complex With 5-Fluorouracil)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: URF, MG, ATP |
 |
Cryo-Em Structure Of Nanodisc-Reconstituted Human Mrp4 Withe1202Q Mutation (In Complex With Lapatinib)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: MEMBRANE PROTEIN Ligands: FMM, MG, ATP |

