Search Count: 24
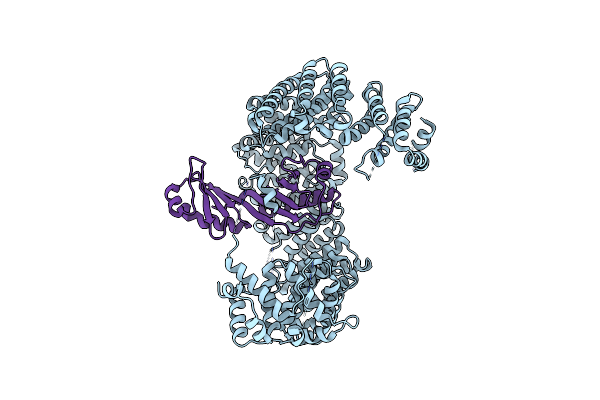 |
The Cryo-Em Structure Of Nuclear Transport Receptor Kap114P Complex With Yeast Tata-Box Binding Protein
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-09-20 Classification: PROTEIN TRANSPORT |
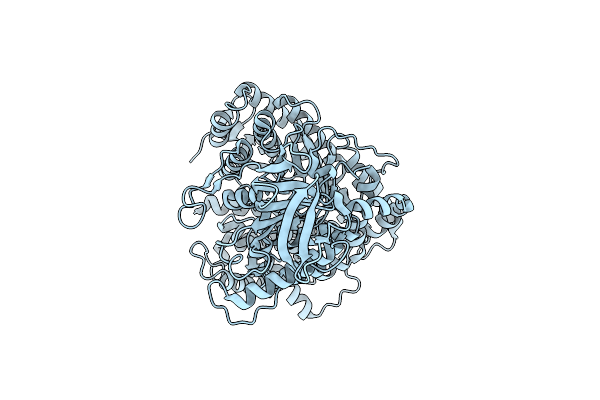 |
Organism: Escherichia coli dh5[alpha]
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: BIOSYNTHETIC PROTEIN |
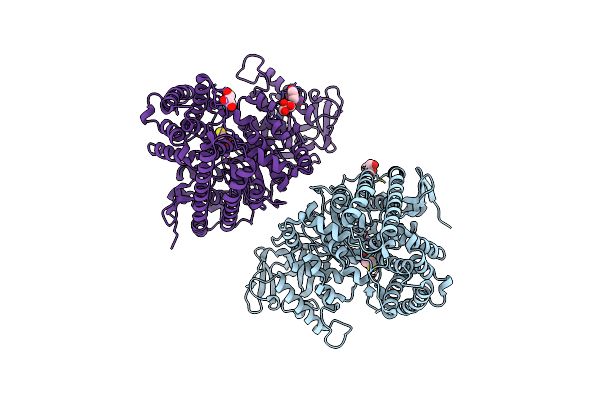 |
Organism: Escherichia coli dh5[alpha]
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-03-29 Classification: BIOSYNTHETIC PROTEIN Ligands: CL, NA, BME, GOL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-02-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2023-02-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
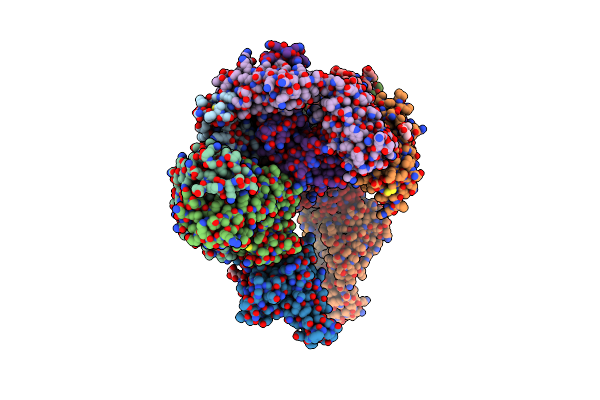 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2023-02-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.37 Å Release Date: 2022-09-28 Classification: IMMUNE SYSTEM |
 |
Cryo-Em Map Of Pedv (Pintung 52) S Protein With All Three Protomers In The D0-Down Conformation Determined In Situ On Intact Viral Particles.
Organism: Porcine epidemic diarrhea virus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Map Of Pedv S Protein With One Protomer In The D0-Up Conformation While The Other Two In The D0-Down Conformation
Organism: Porcine epidemic diarrhea virus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Porcine epidemic diarrhea virus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Map Of Ipec-J2 Cell-Derived Pedv Pt52 S Protein One D0-Down And Two D0-Up
Organism: Porcine epidemic diarrhea virus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Symmetry-Expanded And Locally Refined Protomer Structure Of Ipec-J2 Cell-Derived Pedv Pt52 S With A Ctd-Close Conformation
Organism: Porcine epidemic diarrhea virus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Symmetry-Expanded And Locally Refined Protomer Structure Of Ipec-J2 Cell-Derived Pedv Pt52 S With A Ctd-Open Conformation
Organism: Porcine epidemic diarrhea virus
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2022-08-03 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-23 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With The Monoclonal Antibody M31A7
Organism: Severe acute respiratory syndrome coronavirus 2, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-03-16 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-11-25 Classification: RECOMBINATION/DNA Ligands: CA, ANP |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-11-18 Classification: RECOMBINATION/DNA Ligands: CA, ANP |

