Search Count: 31
 |
Organism: Klebsiella phage vlc6
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: TRS, GOL, BCN |
 |
Crystal Structure Of Trimeric K2-2 Tsp In Complex With Tetrasaccharide And Octasaccharide
Organism: Klebsiella phage vlc6
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: ACE, EDO |
 |
Organism: Klebsiella phage vlc6
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Meiothermus taiwanensis wr-220
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Crystal Structure Of Cbm Deleted Mtglu5 From Meiothermus Taiwanensis Wr-220
Organism: Meiothermus taiwanensis wr-220
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: TRP, MET |
 |
Organism: Meiothermus taiwanensis wr-220
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: BGC, GOL, TRP |
 |
Organism: Meiothermus taiwanensis wr-220
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: GOL, TRP |
 |
Organism: Meiothermus taiwanensis wr-220
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: BGC, SO4 |
 |
Crystal Structure Of Klebsiella Pneumoniae K1 Capsule-Specific Polysaccharide Lyase In A P1 Crystal Form
Organism: Klebsiella phage ntuh-k2044-k1-1
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-05-18 Classification: VIRAL PROTEIN Ligands: LMR, IMD, GOL |
 |
Crystal Structure Of Klebsiella Pneumoniae K1 Capsule-Specific Polysaccharide Lyase In A C2 Crystal Form
Organism: Klebsiella phage ntuh-k2044-k1-1
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2022-05-18 Classification: VIRAL PROTEIN Ligands: CIT, CO3 |
 |
Crystal Structure Of Klebsiella Pneumoniae K1 Capsule-Specific Polysaccharide Lyase In Complex With Products
Organism: Klebsiella phage ntuh-k2044-k1-1
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-05-18 Classification: VIRAL PROTEIN Ligands: IMD, GOL, 98X |
 |
Organism: Meiothermus taiwanensis wr-220
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-10-18 Classification: HYDROLASE Ligands: CA, SO4 |
 |
Organism: Unidentified phage
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2017-03-08 Classification: VIRAL PROTEIN Ligands: MLA |
 |
Crystal Structure Of Phiab6 Tailspike In Complex With Five-Repeated Oligosaccharides Of Acinetobacter Baumannii Surface Polysaccharide
Organism: Unidentified phage
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2017-03-08 Classification: VIRAL PROTEIN Ligands: MLA, ACY |
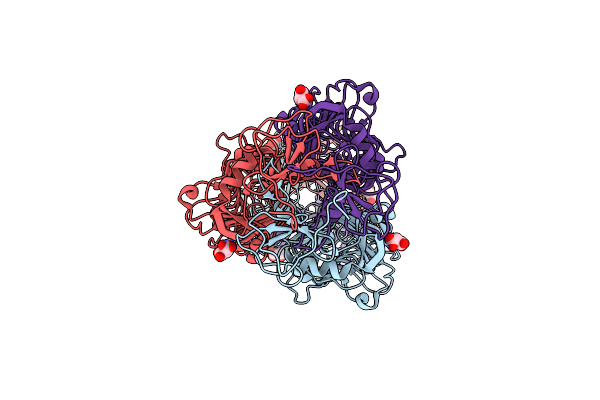 |
Crystal Structure Of Phiab6 Tailspike In Complex With Three-Repeated Oligosaccharides Of Acinetobacter Baumannii Surface Polysaccharide
Organism: Unidentified phage
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2017-03-08 Classification: VIRAL PROTEIN Ligands: MLA, ACY |
 |
Structural Basis Of Dna Recognition By The Effector Domain Of Klebsiella Pneumoniae Pmra
Organism: Klebsiella pneumoniae
Method: SOLUTION NMR Release Date: 2014-01-22 Classification: SIGNALING PROTEIN |
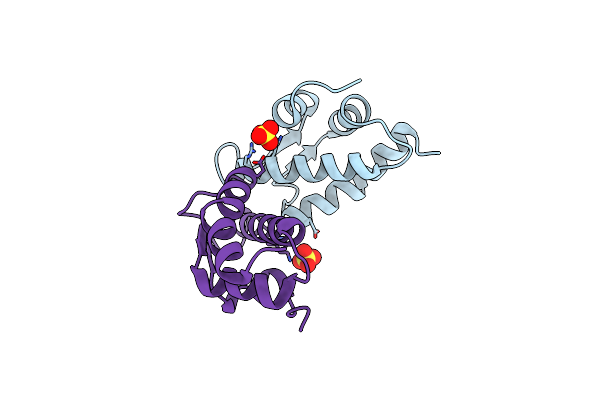 |
Crystal Structure Of Alpha Sub-Domain Of Lon Protease From Brevibacillus Thermoruber
Organism: Brevibacillus thermoruber
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2013-09-11 Classification: HYDROLASE Ligands: SO4 |
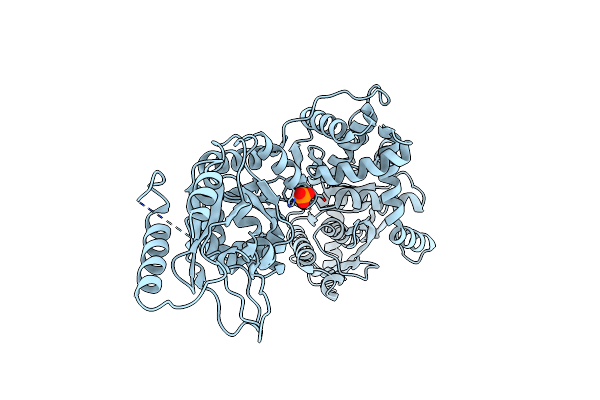 |
Organism: Meiothermus taiwanensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-06-26 Classification: HYDROLASE Ligands: PO4 |
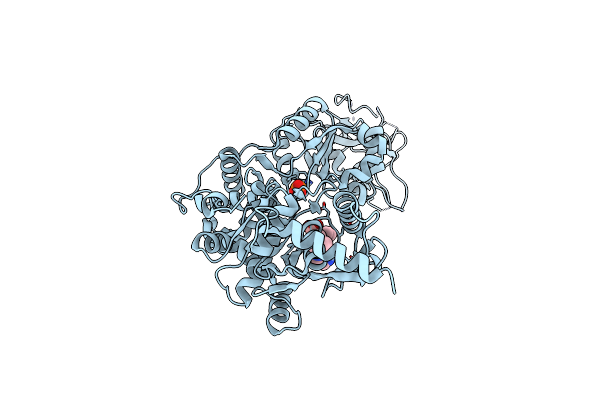 |
Crystal Structure Of The Lon-Like Protease Mtalonc In Complex With Bortezomib
Organism: Meiothermus taiwanensis
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2013-06-26 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: BO2, PO4 |
 |
Crystal Structure Of The Lon-Like Protease Mtalonc In Complex With Lactacystin
Organism: Meiothermus taiwanensis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2013-06-26 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SLA, PO4 |

