Search Count: 530
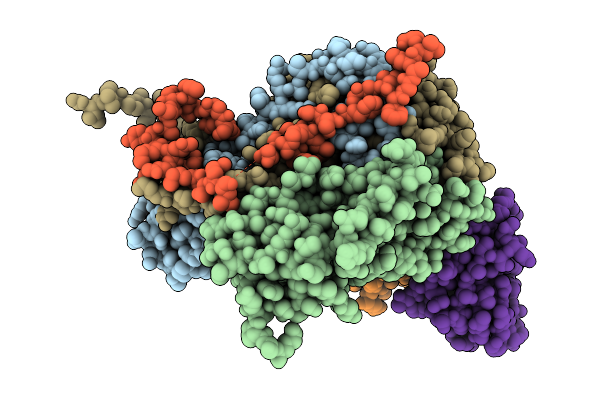 |
Complex Of Fmdv Asia1/Js/05 And Porcine-Derived Neutralizing Monoclonal Antibody Pas12
Organism: Foot-and-mouth disease virus asia 1, Sus scrofa
Method: ELECTRON MICROSCOPY Resolution:2.14 Å Release Date: 2026-01-21 Classification: VIRUS/IMMUNE SYSTEM |
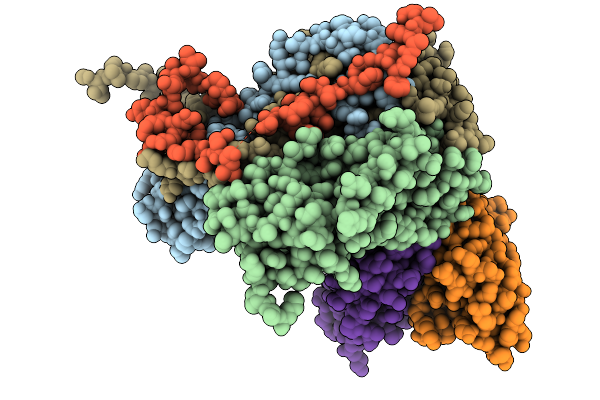 |
Complex Of Fmdv Asia1/Js/05 And Porcine-Derived Neutralizing Monoclonal Antibody Pas5
Organism: Foot-and-mouth disease virus asia 1, Sus scrofa
Method: ELECTRON MICROSCOPY Resolution:2.17 Å Release Date: 2026-01-21 Classification: VIRUS/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
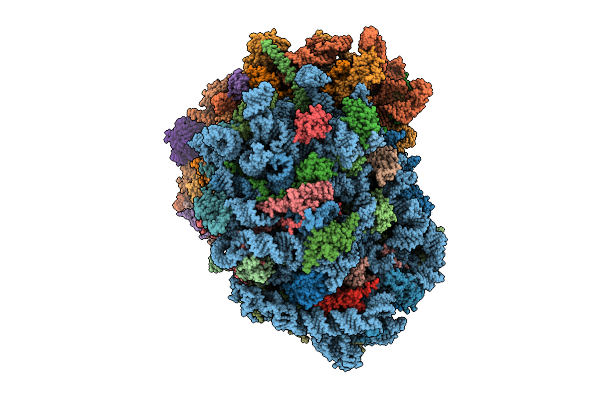 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, ZN, SPM, SPD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
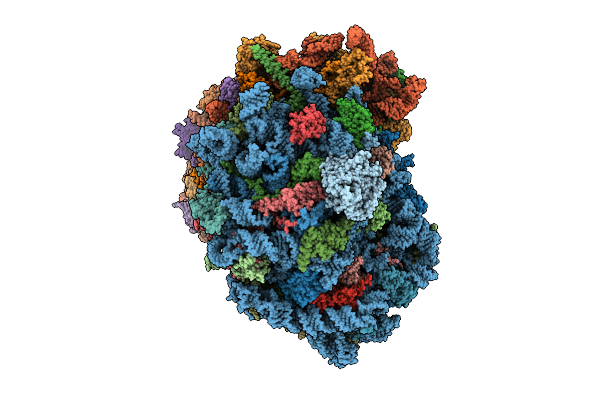 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
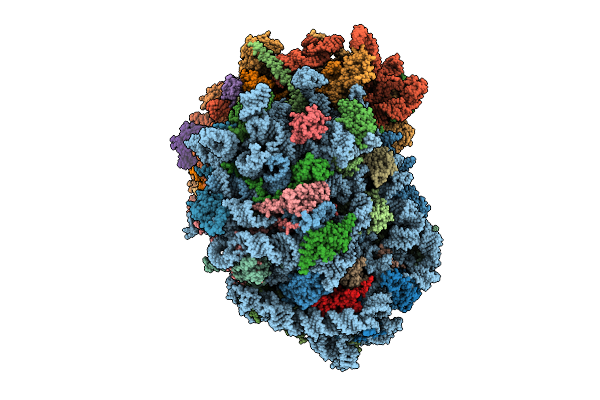 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: ZN, MG, SPM, SPD, 3HE, HMT |
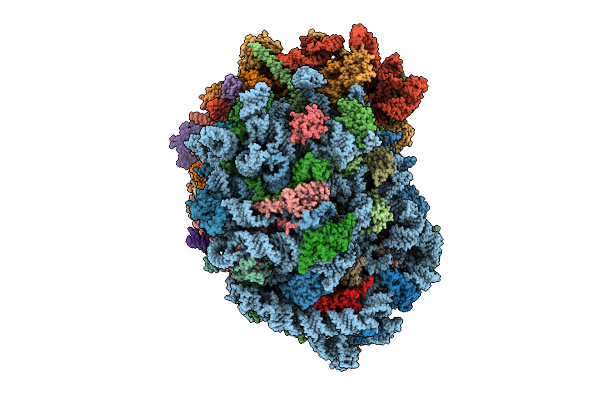 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, ZN, SPM, SPD, 3HE, HMT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, HMT, 3HE, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, 3HE, HMT, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, HMT, 3HE, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RIBOSOME Ligands: MG, SPM, SPD, ZN |

