Search Count: 13
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SUGAR BINDING PROTEIN Ligands: A1IZO, BDF |
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SUGAR BINDING PROTEIN Ligands: BDF, A1IZO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2024-12-18 Classification: HYDROLASE Ligands: GTQ, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-05-18 Classification: OXIDOREDUCTASE Ligands: DHB, NI, ZN, CL |
 |
Crystal Structure Of Histidine-Containing Phosphotransfer Protein B (Hptb) From Pseudomonas Aeruginosa Pao1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-11-04 Classification: TRANSFERASE |
 |
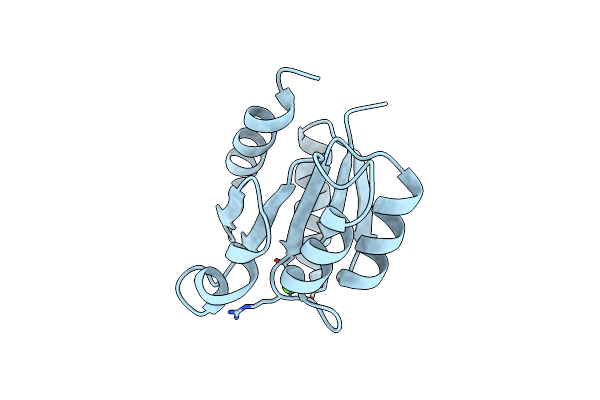
Crystal Structure Of The Receiver Domain Of Sensor Histidine Kinase Pa1611 (Pa1611Rec) From Pseudomonas Aeruginosa Pao1 With Magnesium Ion Coordinated In The Active Site Cleft
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-11-04 Classification: TRANSFERASE Ligands: MG |
 |
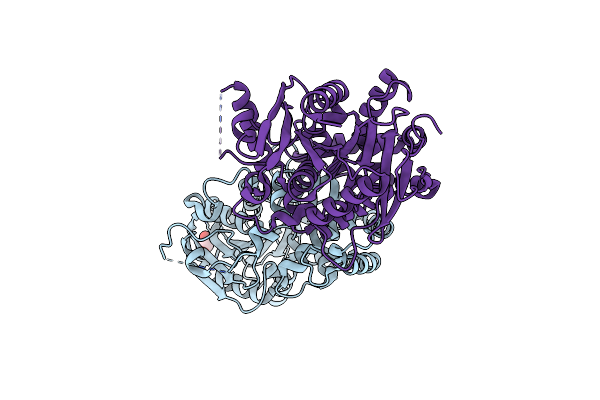
Crystal Structure Of The Receiver Domain Of Sensor Histidine Kinase Pa1611 (Pa1611Rec) From Pseudomonas Aeruginosa Pao1 With Calcium Ion Coordinated In The Active Site Cleft
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2020-11-04 Classification: TRANSFERASE Ligands: CA |
 |
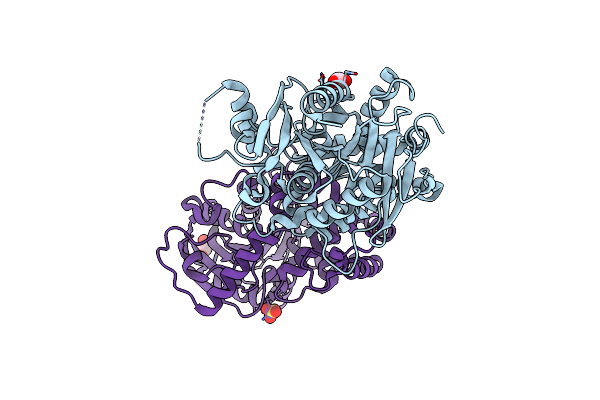
Structure Of The A214C/A250I Mutant Of An Epoxide Hydrolase From Aspergillus Usamii E001 (Aueh2) At 1.48 Angstroms Resolution
Organism: Aspergillus usamii
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2019-12-11 Classification: HYDROLASE Ligands: GOL |
 |
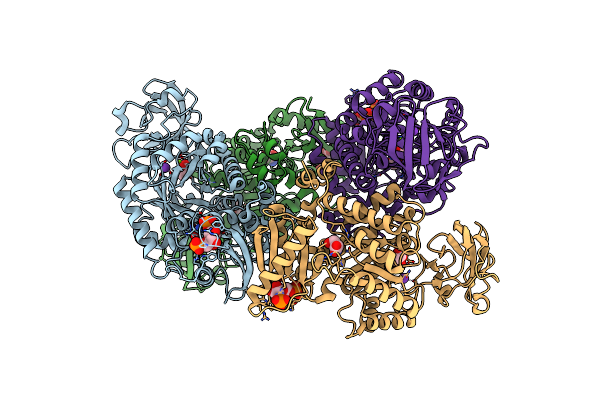
Structure Of An Epoxide Hydrolase From Aspergillus Usamii E001 (Aueh2) At 1.51 Angstroms Resolution
Organism: Aspergillus usamii
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2019-12-11 Classification: HYDROLASE Ligands: ACT, GOL, SO4, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2018-01-31 Classification: TRANSFERASE Ligands: FBP, SER, PYR, MG, K |
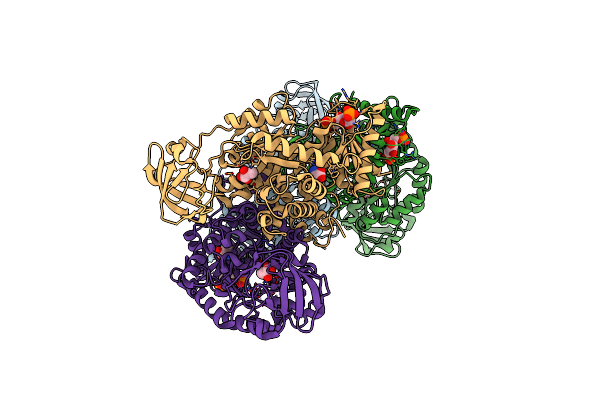 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2016-03-09 Classification: TRANSFERASE Ligands: PYR, MG, SER, FBP |
 |
X-Ray Crystal Structure Of The M6" Riboswitch Aptamer Bound To Pyrimido[4,5-D]Pyrimidine-2,4-Diamine (Ppda)
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2014-07-16 Classification: RNA Ligands: 29G, MG |
 |
X-Ray Crystal Structure Of The M6C" Riboswitch Aptamer Bound To 2-Aminopyrimido[4,5-D]Pyrimidin-4(3H)-One (Ppao)
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-07-16 Classification: RNA Ligands: 29H, MG |

