Search Count: 246
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: DE NOVO PROTEIN Ligands: IOD |
 |
Structural Insights Into Selective Antagonism Of Tg6-129 And Ep4 Prostaglandin Receptor
Organism: Mus musculus, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1EC5 |
 |
Structural Insights Into Selective Antagonism Grapiprant And Ep4 Prostaglandin Receptor
Organism: Mus musculus, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1ECR |
 |
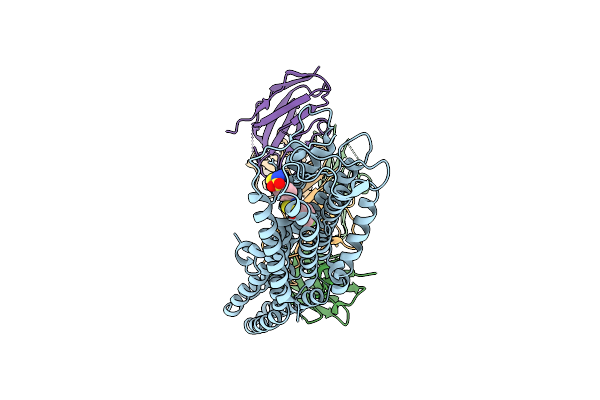
Structural Insights Into Selective Antagonism Of Pf04418948 And Ep2 Prostaglandin Receptor
Organism: Mus musculus, Synthetic construct, Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1ECS |
 |
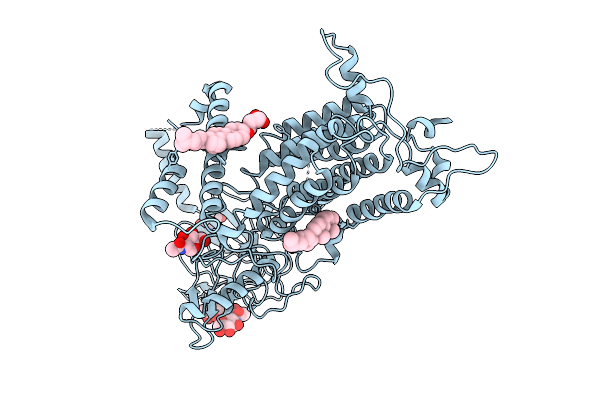
Structural Insights Into Selective Antagonism Of Tg6-129 And Ep2 Prostaglandin Receptor
Organism: Synthetic construct, Homo sapiens, Escherichia coli, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1EC5 |
 |
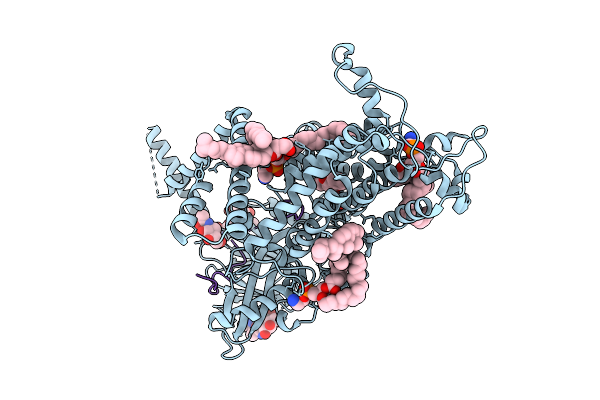
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: NAG, CLR, Y01 |
 |
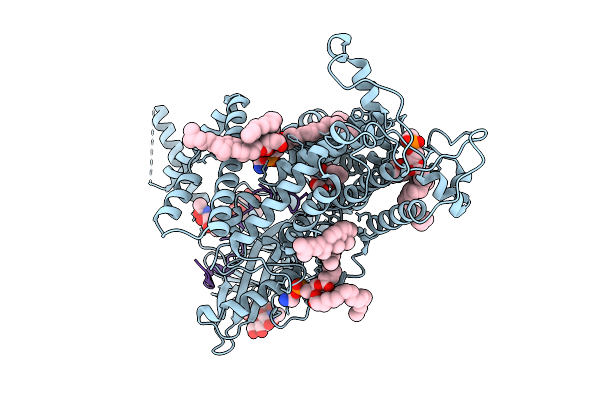
Cryo-Em Structure Of Vitamin K-Dependent Gamma-Carboxylase Complexed With Anisindione
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: NAG, A1AT0, CLR, PEE, Y01, BCT |
 |
Cryo-Em Structure Of Vitamin K-Dependent Gamma-Carboxylase Complexed With Factor Ix
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: NAG, A1AVC, BCT, CO2, CLR, PEE, Y01 |
 |
Cryo-Em Structure Of Vitamin K-Dependent Gamma-Carboxylase Complexed With Osteocalcin
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: NAG, A1AVC, CLR, PEE, Y01, CO2, BCT |
 |
Cryo-Em Structure Of Vitamin K-Dependent Gamma-Carboxylase Complexed With Matrix Gla Protein
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: NAG, A1AVC, CLR, PEE, Y01 |
 |
Cryo-Em Structure Of Vitamin K-Dependent Gamma-Carboxylase Complexed With Factor Ix(Gla)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: NAG, A1AVC, CLR, PEE, Y01, CO2, BCT |
 |
Cryo-Em Structure Of Vitamin K-Dependent Gamma-Carboxylase Complexed With Vitamin K1 2,3-Epoxide
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: NAG, A1EIL, CLR, PEE, Y01, BCT |
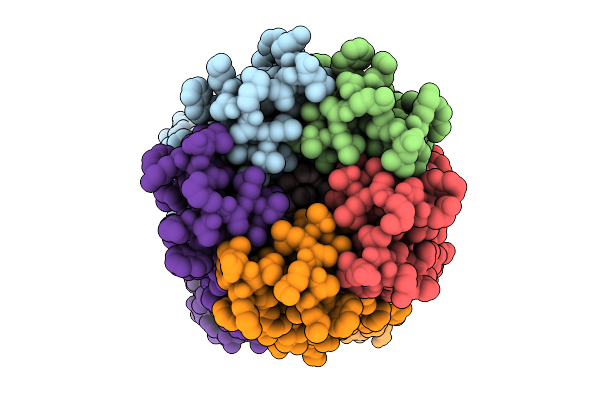 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DE NOVO PROTEIN Ligands: LMT |
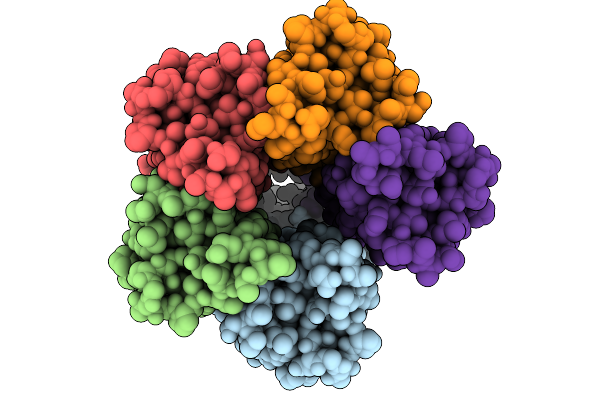 |
Cryo-Em Structure Of A De Novo Designed Voltage-Gated Anion Channel (Dvgac)
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DE NOVO PROTEIN |
 |
Cryo-Em Structure Of Histamine-Bound Histamine Receptor 3 H3R G Protein Complex
Organism: Homo sapiens, Bos taurus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: HSM |
 |
Cryo-Em Structure Of Histamine-Bound Histamine Receptor 4 H4R G Protein Complex
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: HSM, PO4 |
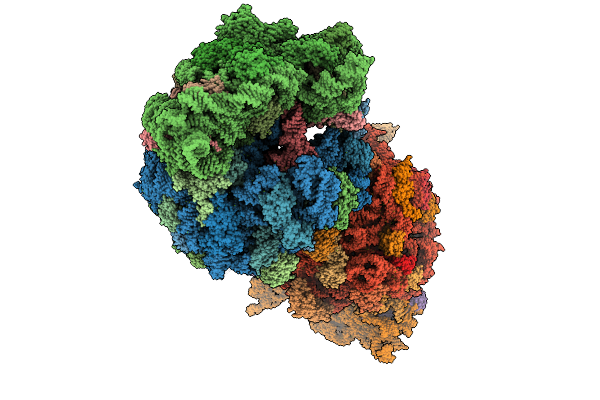 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With O-Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: RIBOSOME Ligands: MG, K, A1A1F, ZN, SF4 |
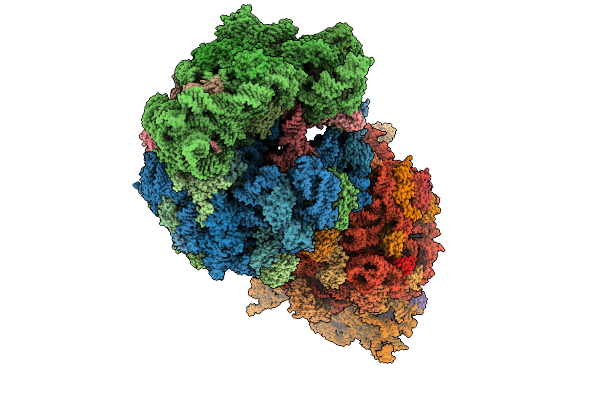 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With C-Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: RIBOSOME Ligands: MG, K, A1A1J, ZN, SF4 |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Se-Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.45A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: RIBOSOME Ligands: MG, K, A1A1K, ZN, SF4 |
 |
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: DE NOVO PROTEIN |

